IL-23 receptor regulates unconventional IL-17-producing T cells that control bacterial infections
- PMID: 20083652
- PMCID: PMC2829977
- DOI: 10.4049/jimmunol.0902796
IL-23 receptor regulates unconventional IL-17-producing T cells that control bacterial infections
Abstract
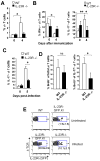
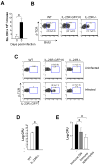
IL-23 plays an important role in autoimmune tissue inflammation and induces the generation of not fully characterized effector cells that mediate protection against pathogens. In this paper, we established the essential role of IL-23R in the host response against intracellular pathogens. IL-23 was critical for the expansion or maintenance of gammadelta and double negative (DN) alphabeta T cells. These cells were rapidly recruited to the site of infection and produced large amounts of IL-17, IFN-gamma, and TNF-alpha. Notably, DN T cells transferred into L. monocytogenes-infected RAG2(-/-) mice prevented bacterial growth, confirming their protective role against intracellular pathogens. Our results show that IL-23 regulates the function of IL-17-producing gammadelta and DN T cells, two essential components of the early protective immune response directed against intracellular pathogens.
Figures





References
-
- Langrish CL, McKenzie BS, Wilson NJ, de Waal Malefyt R, Kastelein RA, Cua DJ. IL-12 and IL-23: master regulators of innate and adaptive immunity. Immunol Rev. 2004;202:96–105. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS045937/NS/NINDS NIH HHS/United States
- R01 NS035685/NS/NINDS NIH HHS/United States
- R01AI073542-01/AI/NIAID NIH HHS/United States
- 2R37NS30843-11/NS/NINDS NIH HHS/United States
- P01 AI056296/AI/NIAID NIH HHS/United States
- R01 AI073542/AI/NIAID NIH HHS/United States
- AI32412/AI/NIAID NIH HHS/United States
- 1P01NS38037-04/NS/NINDS NIH HHS/United States
- 2R01NS35685-06/NS/NINDS NIH HHS/United States
- P01 AI56296/AI/NIAID NIH HHS/United States
- R37 NS030843/NS/NINDS NIH HHS/United States
- 2P01A139671-07/PHS HHS/United States
- P01 AI073748/AI/NIAID NIH HHS/United States
- 1R01A144880-03/PHS HHS/United States
- 1R01NS045937-01/NS/NINDS NIH HHS/United States
- R01 NS046414/NS/NINDS NIH HHS/United States
- 1R01NS046414/NS/NINDS NIH HHS/United States
- P01 NS038037/NS/NINDS NIH HHS/United States
- R01 AI032412/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

