Functional characterization of the Chlamydomonas reinhardtii ERG3 ortholog, a gene involved in the biosynthesis of ergosterol
- PMID: 20084111
- PMCID: PMC2799552
- DOI: 10.1371/journal.pone.0008659
Functional characterization of the Chlamydomonas reinhardtii ERG3 ortholog, a gene involved in the biosynthesis of ergosterol
Erratum in
-
Correction: Functional Characterization of the Chlamydomonas reinhardtii ERG3 Ortholog, a Gene Involved in the Biosynthesis of Ergosterol.PLoS One. 2015 May 28;10(5):e0129189. doi: 10.1371/journal.pone.0129189. eCollection 2015. PLoS One. 2015. PMID: 26020951 Free PMC article. No abstract available.
Abstract
Background: The predominant sterol in the membranes of the alga Chlamydomonas reinhardtii is ergosterol, which is commonly found in the membranes of fungi, but is rarely found in higher plants. Higher plants and fungi synthesize sterols by different pathways, with plants producing cycloartenol as a precursor to end-product sterols, while non-photosynthesizing organisms like yeast and humans produce lanosterol as a precursor. Analysis of the C. reinhardtii genome sequence reveals that this algae is also likely to synthesize sterols using a pathway resembling the higher plant pathway, indicating that its sterols are synthesized somewhat differently than in fungi. The work presented here seeks to establish experimental evidence to support the annotated molecular function of one of the sterol biosynthetic genes in the Chlamydomonas genome.
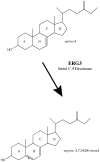
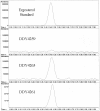
Methodology/principal findings: A gene with homology to the yeast sterol C-5 desaturase, ERG3, is present in the Chlamydomonas genome. To test whether the ERG3 ortholog of C. reinhardtii encodes a sterol C-5 desaturase, Saccharomyces cerevisiae ERG3 knockout strains were created and complemented with a plasmid expressing the Chlamydomonas ERG3. Expression of C. reinhardtii ERG3 cDNA in erg3 null yeast was able to restore ergosterol biosynthesis and reverse phenotypes associated with lack of ERG3 function.
Conclusions/significance: Complementation of the yeast erg3 null phenotypes strongly suggests that the gene annotated as ERG3 in C. reinhardtii functions as a sterol C-5 desaturase.
Conflict of interest statement
Figures



References
-
- Kleinhans FW, Lees ND, Bard M, Haak RA, Woods RA. ESR determinations of membrane permeability in a yeast sterol mutant. Chem Phys Lipids. 1979;23:143–154. - PubMed
-
- Lees ND, Bard M, Kemple MD, Haak RA, Kleinhans FW. ESR determination of membrane order parameter in yeast sterol mutants. Biochim Biophys Acta. 1979;553:469–475. - PubMed
-
- Lees ND, Kleinhans FW, Broughton MC, Pennington DE, Ricker VA, et al. Membrane fluidity alterations in a cytochrome P-450-deficient mutant of Candida albicans. Steroids. 1989;53:567–578. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

