Microphthalmia in Texel sheep is associated with a missense mutation in the paired-like homeodomain 3 (PITX3) gene
- PMID: 20084168
- PMCID: PMC2805710
- DOI: 10.1371/journal.pone.0008689
Microphthalmia in Texel sheep is associated with a missense mutation in the paired-like homeodomain 3 (PITX3) gene
Abstract
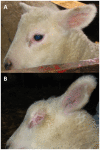
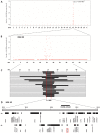
Microphthalmia in sheep is an autosomal recessive inherited congenital anomaly found within the Texel breed. It is characterized by extremely small or absent eyes and affected lambs are absolutely blind. For the first time, we use a genome-wide ovine SNP array for positional cloning of a Mendelian trait in sheep. Genotyping 23 cases and 23 controls using Illumina's OvineSNP50 BeadChip allowed us to localize the causative mutation for microphthalmia to a 2.4 Mb interval on sheep chromosome 22 by association and homozygosity mapping. The PITX3 gene is located within this interval and encodes a homeodomain-containing transcription factor involved in vertebrate lens formation. An abnormal development of the lens vesicle was shown to be the primary event in ovine microphthalmia. Therefore, we considered PITX3 a positional and functional candidate gene. An ovine BAC clone was sequenced, and after full-length cDNA cloning the PITX3 gene was annotated. Here we show that the ovine microphthalmia phenotype is perfectly associated with a missense mutation (c.338G>C, p.R113P) in the evolutionary conserved homeodomain of PITX3. Selection against this candidate causative mutation can now be used to eliminate microphthalmia from Texel sheep in production systems. Furthermore, the identification of a naturally occurring PITX3 mutation offers the opportunity to use the Texel as a genetically characterized large animal model for human microphthalmia.
Conflict of interest statement
Figures

References
-
- Graw J. The genetic and molecular basis of congenital eye defects. Nat Rev Genet. 2003;4:876–888. - PubMed
-
- Chow RL, Lang RA. Early eye development in vertebrates. Annu Rev Cell Dev Biol. 2001;17:255–296. - PubMed
-
- Fantes J, Ragge NK, Lynch SA, McGill NI, Collin JR, et al. Mutations in SOX2 cause anophthalmia. Nat Genet. 2003;33:461–462. - PubMed
-
- Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, et al. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7:463–471. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

