Heterologous expression of wheat VERNALIZATION 2 (TaVRN2) gene in Arabidopsis delays flowering and enhances freezing tolerance
- PMID: 20084169
- PMCID: PMC2805711
- DOI: 10.1371/journal.pone.0008690
Heterologous expression of wheat VERNALIZATION 2 (TaVRN2) gene in Arabidopsis delays flowering and enhances freezing tolerance
Abstract
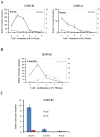
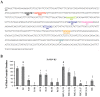
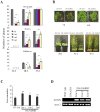
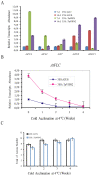
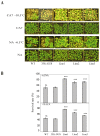
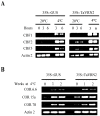
The vernalization gene 2 (VRN2), is a major flowering repressor in temperate cereals that is regulated by low temperature and photoperiod. Here we show that the gene from Triticum aestivum (TaVRN2) is also regulated by salt, heat shock, dehydration, wounding and abscissic acid. Promoter analysis indicates that TaVRN2 regulatory region possesses all the specific responsive elements to these stresses. This suggests pleiotropic effects of TaVRN2 in wheat development and adaptability to the environment. To test if TaVRN2 can act as a flowering repressor in species different from the temperate cereals, the gene was ectopically expressed in the model plant Arabidopsis. Transgenic plants showed no alteration in morphology, but their flowering time was significantly delayed compared to controls plants, indicating that TaVRN2, although having no ortholog in Brassicaceae, can act as a flowering repressor in these species. To identify the possible mechanism by which TaVRN2 gene delays flowering in Arabidopsis, the expression level of several genes involved in flowering time regulation was determined. The analysis indicates that the late flowering of the 35S::TaVRN2 plants was associated with a complex pattern of expression of the major flowering control genes, FCA, FLC, FT, FVE and SOC1. This suggests that heterologous expression of TaVRN2 in Arabidopsis can delay flowering by modulating several floral inductive pathways. Furthermore, transgenic plants showed higher freezing tolerance, likely due to the accumulation of CBF2, CBF3 and the COR genes. Overall, our data suggests that TaVRN2 gene could modulate a common regulator of the two interacting pathways that regulate flowering time and the induction of cold tolerance. The results also demonstrate that TaVRN2 could be used to manipulate flowering time and improve cold tolerance in other species.
Conflict of interest statement
Figures

Similar articles
-
Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering.PLoS Genet. 2012;8(12):e1003134. doi: 10.1371/journal.pgen.1003134. Epub 2012 Dec 13. PLoS Genet. 2012. PMID: 23271982 Free PMC article.
-
TaVRT2 represses transcription of the wheat vernalization gene TaVRN1.Plant J. 2007 Aug;51(4):670-80. doi: 10.1111/j.1365-313X.2007.03172.x. Epub 2007 Jun 22. Plant J. 2007. PMID: 17587304
-
FVE, an Arabidopsis homologue of the retinoblastoma-associated protein that regulates flowering time and cold response, binds to chromatin as a large multiprotein complex.Mol Cells. 2011 Sep;32(3):227-34. doi: 10.1007/s10059-011-1022-6. Epub 2011 Jun 23. Mol Cells. 2011. PMID: 21710206 Free PMC article.
-
The molecular biology of seasonal flowering-responses in Arabidopsis and the cereals.Ann Bot. 2009 Jun;103(8):1165-72. doi: 10.1093/aob/mcp063. Epub 2009 Mar 21. Ann Bot. 2009. PMID: 19304997 Free PMC article. Review.
-
Epigenetic regulation of flowering.Curr Opin Plant Biol. 2007 Oct;10(5):520-7. doi: 10.1016/j.pbi.2007.06.009. Epub 2007 Aug 20. Curr Opin Plant Biol. 2007. PMID: 17709278 Review.
Cited by
-
Genome-Wide Identification and Analysis of the Polycomb Group Family in Medicago truncatula.Int J Mol Sci. 2021 Jul 14;22(14):7537. doi: 10.3390/ijms22147537. Int J Mol Sci. 2021. PMID: 34299158 Free PMC article.
-
The role of DNA content in shaping chromatin architecture and gene expression.Plant J. 2025 Mar;121(6):e70116. doi: 10.1111/tpj.70116. Plant J. 2025. PMID: 40127924 Free PMC article.
-
Transcriptional Regulations on the Low-Temperature-Induced Floral Transition in an Orchidaceae Species, Dendrobium nobile: An Expressed Sequence Tags Analysis.Comp Funct Genomics. 2012;2012:757801. doi: 10.1155/2012/757801. Epub 2012 Apr 9. Comp Funct Genomics. 2012. PMID: 22550428 Free PMC article.
-
Silencing of TaBTF3 gene impairs tolerance to freezing and drought stresses in wheat.Mol Genet Genomics. 2013 Nov;288(11):591-9. doi: 10.1007/s00438-013-0773-5. Epub 2013 Aug 14. Mol Genet Genomics. 2013. PMID: 23942841
-
Transcriptome analysis of an mvp mutant reveals important changes in global gene expression and a role for methyl jasmonate in vernalization and flowering in wheat.J Exp Bot. 2014 Jun;65(9):2271-86. doi: 10.1093/jxb/eru102. Epub 2014 Mar 28. J Exp Bot. 2014. PMID: 24683181 Free PMC article.
References
-
- Henderson IR, Shindo C, Dean C. The need for winter in the switch to flowering. Annu Rev Genet. 2003;37:371–392. - PubMed
-
- Chouard P. Vernalization and its relations to dormancy. Annu Rev Plant Physiol. 1960;11:191–237.
-
- Thomashow MF. Molecular genetics of cold acclimation in higher plants. Adv Gene. 1990;28:99–131.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

