Genetic and molecular analyses of PEG10 reveal new aspects of genomic organization, transcription and translation
- PMID: 20084274
- PMCID: PMC2800197
- DOI: 10.1371/journal.pone.0008686
Genetic and molecular analyses of PEG10 reveal new aspects of genomic organization, transcription and translation
Abstract
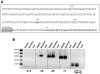
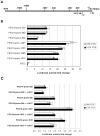
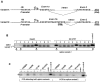
The paternally expressed gene PEG10 is a retrotransposon derived gene adapted through mammalian evolution located on human chromosome 7q21. PEG10 codes for at least two proteins, PEG10-RF1 and PEG10-RF1/2, by -1 frameshift translation. Overexpression or reinduced PEG10 expression was seen in malignancies, like hepatocellular carcinoma or B-cell acute and chronic lymphocytic leukemia. PEG10 was also shown to promote adipocyte differentiation. Experimental evidence suggests that the PEG10-RF1 protein is an inhibitor of apoptosis and mediates cell proliferation. Here we present new data on the genomic organization of PEG10 by identifying the major transcription start site, a new splice variant and report the cloning and analysis of 1.9 kb of the PEG10 promoter. Furthermore, we show for the first time that PEG10 translation is initiated at a non-AUG start codon upstream of the previously predicted AUG codon as well as at the AUG codon. The finding that PEG10 translation is initiated at different sides adds a new aspect to the already interesting feature of PEG10's -1 frameshift translation mechanism. It is now important to unravel the cellular functions of the PEG10 protein variants and how they are related to normal or pathological conditions. The generated promoter-reporter constructs can be used for future studies to investigate how PEG10 expression is regulated. In summary, our study provides new data on the genomic organization as well as expression and translation of PEG10, a prerequisite in order to study and understand the role of PEG10 in cancer, embryonic development and normal cell homeostasis.
Conflict of interest statement
Figures





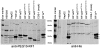
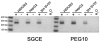
Similar articles
-
Mammalian gene PEG10 expresses two reading frames by high efficiency -1 frameshifting in embryonic-associated tissues.J Biol Chem. 2007 Dec 28;282(52):37359-69. doi: 10.1074/jbc.M705676200. Epub 2007 Oct 16. J Biol Chem. 2007. PMID: 17942406
-
Functional Study of the Retrotransposon-Derived Human PEG10 Protease.Int J Mol Sci. 2020 Mar 31;21(7):2424. doi: 10.3390/ijms21072424. Int J Mol Sci. 2020. PMID: 32244497 Free PMC article.
-
Human retroviral gag- and gag-pol-like proteins interact with the transforming growth factor-beta receptor activin receptor-like kinase 1.J Biol Chem. 2005 Mar 4;280(9):8482-93. doi: 10.1074/jbc.M409197200. Epub 2004 Dec 16. J Biol Chem. 2005. PMID: 15611116
-
Roles of PEG10 in cancer and neurodegenerative disorder (Review).Oncol Rep. 2025 May;53(5):60. doi: 10.3892/or.2025.8893. Epub 2025 Apr 4. Oncol Rep. 2025. PMID: 40183369 Free PMC article. Review.
-
Prognostic value of PEG10 in Asian solid tumors: A meta-analysis.Clin Chim Acta. 2018 Aug;483:197-203. doi: 10.1016/j.cca.2018.04.041. Epub 2018 May 1. Clin Chim Acta. 2018. PMID: 29727698 Review.
Cited by
-
UBQLN2 restrains the domesticated retrotransposon PEG10 to maintain neuronal health in ALS.Elife. 2023 Mar 23;12:e79452. doi: 10.7554/eLife.79452. Elife. 2023. PMID: 36951542 Free PMC article.
-
PEG10 as an oncogene: expression regulatory mechanisms and role in tumor progression.Cancer Cell Int. 2018 Aug 13;18:112. doi: 10.1186/s12935-018-0610-3. eCollection 2018. Cancer Cell Int. 2018. PMID: 30123090 Free PMC article. Review.
-
Genetic variations in plasma circulating DNA of HBV-related hepatocellular carcinoma patients predict recurrence after liver transplantation.PLoS One. 2011;6(10):e26003. doi: 10.1371/journal.pone.0026003. Epub 2011 Oct 5. PLoS One. 2011. Retraction in: PLoS One. 2019 Sep 4;14(9):e0222258. doi: 10.1371/journal.pone.0222258. PMID: 21998744 Free PMC article. Retracted.
-
Human Virus-Like Proteins: Implications for Gene Therapy.Curr Gene Ther. 2025;25(3):227-236. doi: 10.2174/0115665232303436240515071754. Curr Gene Ther. 2025. PMID: 38798208 Review.
-
Control of gene expression by translational recoding.Adv Protein Chem Struct Biol. 2012;86:129-49. doi: 10.1016/B978-0-12-386497-0.00004-9. Adv Protein Chem Struct Biol. 2012. PMID: 22243583 Free PMC article. Review.
References
-
- Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. - PubMed
-
- Lux A, Beil C, Majety M, Barron S, Gallione CJ, et al. Human retroviral gag- and gag-pol-like proteins interact with the transforming growth factor-beta receptor activin receptor-like kinase 1. J Biol Chem. 2005;280:8482–8493. - PubMed
-
- Wills NM, Moore B, Hammer A, Gesteland RF, Atkins JF. A functional -1 ribosomal frameshift signal in the human paraneoplastic Ma3 gene. J Biol Chem. 2006;281:7082–7088. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

