Engineering fluorometabolite production: fluorinase expression in Salinispora tropica Yields Fluorosalinosporamide
- PMID: 20085308
- PMCID: PMC2846182
- DOI: 10.1021/np900719u
Engineering fluorometabolite production: fluorinase expression in Salinispora tropica Yields Fluorosalinosporamide
Abstract
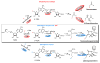
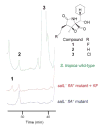
Organofluorine compounds play an important role in medicinal chemistry, where they are responsible for up to 15% of the pharmaceutical products on the market. While natural products are valuable sources of new chemical entities, natural fluorinated molecules are extremely rare and the pharmaceutical industry has not benefited from a microbial source of this class of compounds. Streptomyces cattleya is an unusual bacterium in that it elaborates fluoroacetate and the amino acid 4-fluorothreonine. The discovery in 2002 of the fluorination enzyme FlA responsible for C-F bond formation in S. cattleya, and its subsequent characterization, opened up for the first time the prospect of genetically engineering fluorometabolite production from fluoride ion in host organisms. As a proof of principle, we report here the induced production of fluorosalinosporamide by replacing the chlorinase gene salL from Salinispora tropica with the fluorinase gene flA.
Figures

References
-
- Harper DB, O'Hagan D. Nat Prod Rep. 1994;11:123–133. - PubMed
-
- Neumann CS, Fujimori DG, Walsh CT. Chem Biol. 2008;15:99–109. - PubMed
-
- Dong C, Huang F, Deng H, Schaffrath C, Spencer JB, O'Hagan D, Naismith JH. Nature. 2004;427:561–565. - PubMed
-
- O'Hagan D, Schaffrath C, Cobb SL, Hamilton JT, Murphy CD. Nature. 2002;416:279. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

