Identification of inhibitors of Plasmodium falciparum phosphoethanolamine methyltransferase using an enzyme-coupled transmethylation assay
- PMID: 20085640
- PMCID: PMC2824672
- DOI: 10.1186/1471-2091-11-4
Identification of inhibitors of Plasmodium falciparum phosphoethanolamine methyltransferase using an enzyme-coupled transmethylation assay
Abstract
Background: The phosphoethanolamine methyltransferase, PfPMT, of the human malaria parasite Plasmodium falciparum, a member of a newly identified family of phosphoethanolamine methyltransferases (PMT) found solely in some protozoa, nematodes, frogs, and plants, is involved in the synthesis of the major membrane phospholipid, phosphatidylcholine. PMT enzymes catalyze a three-step S-adenosylmethionine-dependent methylation of the nitrogen atom of phosphoethanolamine to form phosphocholine. In P. falciparum, this activity is a limiting step in the pathway of synthesis of phosphatidylcholine from serine and plays an important role in the development, replication and survival of the parasite within human red blood cells.
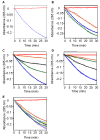
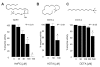
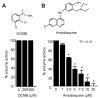
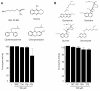
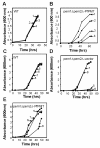
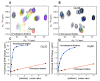
Results: We have employed an enzyme-coupled methylation assay to screen for potential inhibitors of PfPMT. In addition to hexadecyltrimethylammonium, previously known to inhibit PfPMT, two compounds dodecyltrimethylammonium and amodiaquine were also found to inhibit PfPMT activity in vitro. Interestingly, PfPMT activity was not inhibited by the amodiaquine analog, chloroquine, or other aminoquinolines, amino alcohols, or histamine methyltransferase inhibitors. Using yeast as a surrogate system we found that unlike wild-type cells, yeast mutants that rely on PfPMT for survival were sensitive to amodiaquine, and their phosphatidylcholine biosynthesis was inhibited by this compound. Furthermore NMR titration studies to characterize the interaction between amoidaquine and PfPMT demonstrated a specific and concentration dependent binding of the compound to the enzyme.
Conclusion: The identification of amodiaquine as an inhibitor of PfPMT in vitro and in yeast, and the biophysical evidence for the specific interaction of the compound with the enzyme will set the stage for the development of analogs of this drug that specifically inhibit this enzyme and possibly other PMTs.
Figures


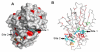
References
-
- WHO. World Malaria Report 2008. Geneva: World Health Organisation; 2008.
-
- Pessi G, Ben Mamoun C. Pathways for phosphatidylcholine biosynthesis: targets and strategies for antimalarial drugs. Future Medicine, Future Lipidology. 2006;1(2):173–180. doi: 10.2217/17460875.1.2.173. - DOI
-
- Vial HJ, Ben Mamoun C, Sherman IW. Molecular Approaches to Malaria. Washington D.C.: ASM Press; 2005. Plasmodium lipids: Metabolism and Function; pp. 327–352.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

