Identification of interacting transcription factors regulating tissue gene expression in human
- PMID: 20085649
- PMCID: PMC2822763
- DOI: 10.1186/1471-2164-11-49
Identification of interacting transcription factors regulating tissue gene expression in human
Abstract
Background: Tissue gene expression is generally regulated by multiple transcription factors (TFs). A major first step toward understanding how tissues achieve their specificity is to identify, at the genome scale, interacting TFs regulating gene expression in different tissues. Despite previous discoveries, the mechanisms that control tissue gene expression are not fully understood.
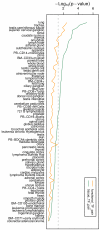
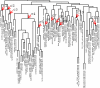
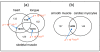
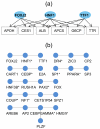
Results: We have integrated a function conservation approach, which is based on evolutionary conservation of biological function, and genes with highest expression level in human tissues to predict TF pairs controlling tissue gene expression. To this end, we have identified 2549 TF pairs associated with a certain tissue. To find interacting TFs controlling tissue gene expression in a broad spatial and temporal manner, we looked for TF pairs common to the same type of tissues and identified 379 such TF pairs, based on which TF-TF interaction networks were further built. We also found that tissue-specific TFs may play an important role in recruiting non-tissue-specific TFs to the TF-TF interaction network, offering the potential for coordinating and controlling tissue gene expression across a variety of conditions.
Conclusion: The findings from this study indicate that tissue gene expression is regulated by large sets of interacting TFs either on the same promoter of a gene or through TF-TF interaction networks.
Figures


Similar articles
-
Computational analysis of tissue-specific combinatorial gene regulation: predicting interaction between transcription factors in human tissues.Nucleic Acids Res. 2006;34(17):4925-36. doi: 10.1093/nar/gkl595. Epub 2006 Sep 18. Nucleic Acids Res. 2006. PMID: 16982645 Free PMC article.
-
Detection of interacting transcription factors in human tissues using predicted DNA binding affinity.BMC Genomics. 2012;13 Suppl 1(Suppl 1):S2. doi: 10.1186/1471-2164-13-S1-S2. Epub 2012 Jan 17. BMC Genomics. 2012. PMID: 22369666 Free PMC article.
-
Inter- and intra-combinatorial regulation by transcription factors and microRNAs.BMC Genomics. 2007 Oct 30;8:396. doi: 10.1186/1471-2164-8-396. BMC Genomics. 2007. PMID: 17971223 Free PMC article.
-
Global analysis of gene transcription regulation in prokaryotes.Cell Mol Life Sci. 2006 Oct;63(19-20):2260-90. doi: 10.1007/s00018-006-6184-6. Cell Mol Life Sci. 2006. PMID: 16927028 Free PMC article. Review.
-
How do you find transcription factors? Computational approaches to compile and annotate repertoires of regulators for any genome.Methods Mol Biol. 2012;786:3-19. doi: 10.1007/978-1-61779-292-2_1. Methods Mol Biol. 2012. PMID: 21938617 Review.
Cited by
-
Genome-wide patterns of promoter sharing and co-expression in bovine skeletal muscle.BMC Genomics. 2011 Jan 12;12:23. doi: 10.1186/1471-2164-12-23. BMC Genomics. 2011. PMID: 21226902 Free PMC article.
-
Interactions Among Plant Transcription Factors Regulating Expression of Stress-responsive Genes.Bioinform Biol Insights. 2014 Sep 10;8:193-8. doi: 10.4137/BBI.S16313. eCollection 2014. Bioinform Biol Insights. 2014. PMID: 25249757 Free PMC article.
-
Detecting Cooperativity between Transcription Factors Based on Functional Coherence and Similarity of Their Target Gene Sets.PLoS One. 2016 Sep 13;11(9):e0162931. doi: 10.1371/journal.pone.0162931. eCollection 2016. PLoS One. 2016. PMID: 27623007 Free PMC article.
-
Long-Term Oral Administration of LLHK, LHK, and HK Alters Gene Expression Profile and Restores Age-Dependent Atrophy and Dysfunction of Rat Salivary Glands.Biomedicines. 2020 Feb 20;8(2):38. doi: 10.3390/biomedicines8020038. Biomedicines. 2020. PMID: 32093221 Free PMC article.
-
coTRaCTE predicts co-occurring transcription factors within cell-type specific enhancers.PLoS Comput Biol. 2018 Aug 24;14(8):e1006372. doi: 10.1371/journal.pcbi.1006372. eCollection 2018 Aug. PLoS Comput Biol. 2018. PMID: 30142147 Free PMC article.
References
-
- Arnone MI, Davidson EH. The hardwiring of development: organization and function of genomic regulatory systems. Development (Cambridge, England) 1997;124(10):1851–1864. - PubMed
-
- Metzger S, Halaas JL, Breslow JL, Sladek FM. Orphan receptor HNF-4 and bZip protein C/EBP alpha bind to overlapping regions of the apolipoprotein B gene promoter and synergistically activate transcription. The Journal of biological chemistry. 1993;268(22):16831–16838. - PubMed
-
- Rada-Iglesias A, Wallerman O, Koch C, Ameur A, Enroth S, Clelland G, Wester K, Wilcox S, Dovey OM, Ellis PD. et al.Binding sites for metabolic disease related transcription factors inferred at base pair resolution by chromatin immunoprecipitation and genomic microarrays. Human molecular genetics. 2005;14(22):3435–3447. doi: 10.1093/hmg/ddi378. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

