Molecular characterization of the virulent infectious hematopoietic necrosis virus (IHNV) strain 220-90
- PMID: 20085652
- PMCID: PMC2820013
- DOI: 10.1186/1743-422X-7-10
Molecular characterization of the virulent infectious hematopoietic necrosis virus (IHNV) strain 220-90
Abstract
Background: Infectious hematopoietic necrosis virus (IHNV) is the type species of the genus Novirhabdovirus, within the family Rhabdoviridae, infecting several species of wild and hatchery reared salmonids. Similar to other rhabdoviruses, IHNV has a linear single-stranded, negative-sense RNA genome of approximately 11,000 nucleotides. The IHNV genome encodes six genes; the nucleocapsid, phosphoprotein, matrix protein, glycoprotein, non-virion protein and polymerase protein genes, respectively. This study describes molecular characterization of the virulent IHNV strain 220-90, belonging to the M genogroup, and its phylogenetic relationships with available sequences of IHNV isolates worldwide.
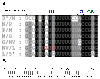
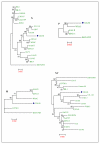
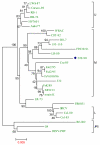
Results: The complete genomic sequence of IHNV strain 220-90 was determined from the DNA of six overlapping clones obtained by RT-PCR amplification of genomic RNA. The complete genome sequence of 220-90 comprises 11,133 nucleotides (GenBank GQ413939) with the gene order of 3'-N-P-M-G-NV-L-5'. These genes are separated by conserved gene junctions, with di-nucleotide gene spacers. An additional uracil nucleotide was found at the end of the 5'-trailer region, which was not reported before in other IHNV strains. The first 15 of the 16 nucleotides at the 3'- and 5'-termini of the genome are complementary, and the first 4 nucleotides at 3'-ends of the IHNV are identical to other novirhadoviruses. Sequence homology and phylogenetic analysis of the glycoprotein genes show that 220-90 strain is 97% identical to most of the IHNV strains. Comparison of the virulent 220-90 genomic sequences with less virulent WRAC isolate shows more than 300 nucleotides changes in the genome, which doesn't allow one to speculate putative residues involved in the virulence of IHNV.
Conclusion: We have molecularly characterized one of the well studied IHNV isolates, 220-90 of genogroup M, which is virulent for rainbow trout, and compared phylogenetic relationship with North American and other strains. Determination of the complete nucleotide sequence is essential for future studies on pathogenesis of IHNV using a reverse genetics approach and developing efficient control strategies.
Figures


Similar articles
-
Phylogenetic relationships of Iranian infectious hematopoietic necrosis virus of rainbow trout (Oncorhynchus mykiss) based on the glycoprotein gene.Arch Virol. 2016 Mar;161(3):657-63. doi: 10.1007/s00705-015-2684-8. Epub 2015 Nov 24. Arch Virol. 2016. PMID: 26602428
-
Determination of the complete genome sequence of infectious hematopoietic necrosis virus (IHNV) Ch20101008 and viral molecular evolution in China.Infect Genet Evol. 2014 Oct;27:418-31. doi: 10.1016/j.meegid.2014.08.013. Epub 2014 Aug 27. Infect Genet Evol. 2014. PMID: 25172153
-
Heterologous Exchanges of Glycoprotein and Non-Virion Protein in Novirhabdoviruses: Assessment of Virulence in Yellow Perch (Perca flavescens) and Rainbow Trout (Oncorhynchus mykiss).Viruses. 2024 Apr 22;16(4):652. doi: 10.3390/v16040652. Viruses. 2024. PMID: 38675990 Free PMC article.
-
Molecular characterization of the Great Lakes viral hemorrhagic septicemia virus (VHSV) isolate from USA.Virol J. 2009 Oct 25;6:171. doi: 10.1186/1743-422X-6-171. Virol J. 2009. PMID: 19852863 Free PMC article.
-
Epidemiological characteristics of infectious hematopoietic necrosis virus (IHNV): a review.Vet Res. 2016 Jun 10;47(1):63. doi: 10.1186/s13567-016-0341-1. Vet Res. 2016. PMID: 27287024 Free PMC article. Review.
Cited by
-
In vivo fitness associated with high virulence in a vertebrate virus is a complex trait regulated by host entry, replication, and shedding.J Virol. 2011 Apr;85(8):3959-67. doi: 10.1128/JVI.01891-10. Epub 2011 Feb 9. J Virol. 2011. PMID: 21307204 Free PMC article.
-
Clathrin-mediated endocytosis in living host cells visualized through quantum dot labeling of infectious hematopoietic necrosis virus.J Virol. 2011 Jul;85(13):6252-62. doi: 10.1128/JVI.00109-11. Epub 2011 Apr 27. J Virol. 2011. PMID: 21525360 Free PMC article.
-
The zebrafish galectins Drgal1-L2 and Drgal3-L1 bind in vitro to the infectious hematopoietic necrosis virus (IHNV) glycoprotein and reduce viral adhesion to fish epithelial cells.Dev Comp Immunol. 2016 Feb;55:241-252. doi: 10.1016/j.dci.2015.09.007. Epub 2015 Sep 30. Dev Comp Immunol. 2016. PMID: 26429411 Free PMC article.
-
Vaccination Route Determines the Kinetics and Magnitude of Nasal Innate Immune Responses in Rainbow Trout (Oncorhynchus mykiss).Biology (Basel). 2020 Oct 1;9(10):319. doi: 10.3390/biology9100319. Biology (Basel). 2020. PMID: 33019693 Free PMC article.
-
Analysis of the genome sequence of infectious hematopoietic necrosis virus HLJ-09 in China.Virus Genes. 2016 Feb;52(1):29-37. doi: 10.1007/s11262-015-1263-0. Epub 2016 Jan 22. Virus Genes. 2016. PMID: 26801781
References
-
- Wolf K. Infectious hematopoietic necrosis virus. In Fish Viruses and Fish Viral Diseases. Ithaca, NY, Cornell University Press; 1988. pp. 83–114.
-
- Winton JR. Recent advances in detection and control of infectious hematopoietic necrosis virus in aquaculture. Annual Rev Fish Dis. 1991;1:83–93. doi: 10.1016/0959-8030(91)90024-E. - DOI
-
- Rucker RR, Whippie WJ, Parvin JR, Evans CA. A contagious disease of salmon possibly of virus origin. US Fish Wildl Serv and Fish Bull. 1953;54:174–175.
-
- Guenther RW, Watson SW, Rucker RR. Etiology of sockeye salmon "virus" disease. US Fish Wildl Serv Spec Sci Rep Fish. 1959;296:1–10.
-
- Wingfield WH, Fryer JL, Pilcher KS. Properties of sockeye salmon virus (Oregon strain) Proc Soc Exp Biol Med. 1969;30:1055–1059. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

