Pannexin1 and pannexin3 delivery, cell surface dynamics, and cytoskeletal interactions
- PMID: 20086016
- PMCID: PMC2838334
- DOI: 10.1074/jbc.M109.082008
Pannexin1 and pannexin3 delivery, cell surface dynamics, and cytoskeletal interactions
Abstract
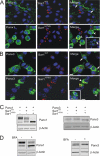
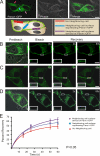
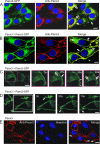
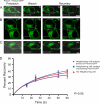
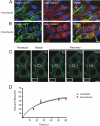
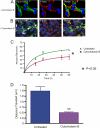
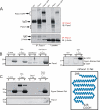
Pannexins (Panx) are a class of integral membrane proteins that have been proposed to exhibit characteristics similar to those of connexin family members. In this study, we utilized Cx43-positive BICR-M1R(k) cells to stably express Panx1, Panx3, or Panx1-green fluorescent protein (GFP) to assess their trafficking, cell surface dynamics, and interplay with the cytoskeletal network. Expression of a Sar1 dominant negative mutant revealed that endoplasmic reticulum to Golgi transport of Panx1 and Panx3 was mediated via COPII-dependent vesicles. Distinct from Cx43-GFP, fluorescence recovery after photobleaching studies revealed that both Panx1-GFP and Panx3-GFP remained highly mobile at the cell surface. Unlike Cx43, Panx1-GFP exhibited no detectable interrelationship with microtubules. Conversely, cytochalasin B-induced disruption of microfilaments caused a severe loss of cell surface Panx1-GFP, a reduction in the recoverable fraction of Panx1-GFP that remained at the cell surface, and a decrease in Panx1-GFP vesicular transport. Furthermore, co-immunoprecipitation and co-sedimentation assays revealed actin as a novel binding partner of Panx1. Collectively, we conclude that although Panx1 and Panx3 share a common endoplasmic reticulum to Golgi secretory pathway to Cx43, their ultimate cell surface residency appears to be independent of cell contacts and the need for intact microtubules. Importantly, Panx1 has an interaction with actin microfilaments that regulates its cell surface localization and mobility.
Figures


References
-
- Panchin Y., Kelmanson I., Matz M., Lukyanov K., Usman N., Lukyanov S. (2000) Curr. Biol. 10, R473–R474 - PubMed
-
- Willecke K., Eiberger J., Degen J., Eckardt D., Romualdi A., Güldenagel M., Deutsch U., Söhl G. (2002) Biol. Chem. 383, 725–737 - PubMed
-
- Baranova A., Ivanov D., Petrash N., Pestova A., Skoblov M., Kelmanson I., Shagin D., Nazarenko S., Geraymovych E., Litvin O., Tiunova A., Born T. L., Usman N., Staroverov D., Lukyanov S., Panchin Y. (2004) Genomics 83, 706–716 - PubMed
-
- Boassa D., Ambrosi C., Qiu F., Dahl G., Gaietta G., Sosinsky G. (2007) J. Biol. Chem. 282, 31733–31743 - PubMed
-
- Penuela S., Bhalla R., Gong X. Q., Cowan K. N., Celetti S. J., Cowan B. J., Bai D., Shao Q., Laird D. W. (2007) J. Cell Sci. 120, 3772–3783 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

