Molecular mechanisms that funnel RNA precursors into endogenous small-interfering RNA and microRNA biogenesis pathways in Drosophila
- PMID: 20086050
- PMCID: PMC2822916
- DOI: 10.1261/rna.1952110
Molecular mechanisms that funnel RNA precursors into endogenous small-interfering RNA and microRNA biogenesis pathways in Drosophila
Abstract
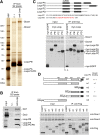
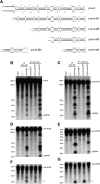
In Drosophila, three types of endogenous small RNAs-microRNAs (miRNAs), PIWI-interacting RNAs (piRNAs), and endogenous small-interfering RNAs (endo-siRNAs or esiRNAs)-function as triggers in RNA silencing. Although piRNAs are produced independently of Dicer, miRNA and esiRNA biogenesis pathways require Dicer1 and Dicer2, respectively. Recent studies have shown that among the four isoforms of Loquacious (Loqs), Loqs-PB and Loqs-PD are involved in miRNA and esiRNA processing pathways, respectively. However, how these Loqs isoforms function in their respective small RNA biogenesis pathways remains elusive. Here, we show that Loqs-PD associates specifically with Dicer2 through its C-terminal domain. The Dicer2-Loqs-PD complex contains R2D2, another known Dicer2 partner, and excises both exogenous siRNAs and esiRNAs from their corresponding precursors in vitro. However, Loqs-PD, but not R2D2, enhanced Dicer2 activity. The Dicer2-Loqs-PD complex processes esiRNA precursor hairpins with long stems, which results in the production of AGO2-associated small RNAs. Interestingly, however, small RNAs derived from terminal hairpins of esiRNA precursors are loaded onto AGO1; thus, they are classified as a new subset of miRNAs. These results suggest that the precursor RNA structure determines the biogenesis mechanism of esiRNAs and miRNAs, thereby implicating hairpin structures with long stems as intermediates in the evolution of Drosophila miRNA.
Figures




Similar articles
-
Loqs-PD and R2D2 define independent pathways for RISC generation in Drosophila.Nucleic Acids Res. 2011 May;39(9):3836-51. doi: 10.1093/nar/gkq1324. Epub 2011 Jan 17. Nucleic Acids Res. 2011. PMID: 21245036 Free PMC article.
-
Structural basis of endo-siRNA processing by Drosophila Dicer-2 and Loqs-PD.Nucleic Acids Res. 2025 Feb 8;53(4):gkaf102. doi: 10.1093/nar/gkaf102. Nucleic Acids Res. 2025. PMID: 39988314 Free PMC article.
-
Processing of Drosophila endo-siRNAs depends on a specific Loquacious isoform.RNA. 2009 Oct;15(10):1886-95. doi: 10.1261/rna.1611309. Epub 2009 Jul 27. RNA. 2009. PMID: 19635780 Free PMC article.
-
Biogenesis of small RNAs in animals.Nat Rev Mol Cell Biol. 2009 Feb;10(2):126-39. doi: 10.1038/nrm2632. Nat Rev Mol Cell Biol. 2009. PMID: 19165215 Review.
-
MicroRNAs: Loquacious speaks out.Curr Biol. 2005 Aug 9;15(15):R603-5. doi: 10.1016/j.cub.2005.07.044. Curr Biol. 2005. PMID: 16085484 Review.
Cited by
-
Drosophila dicer-2 cleavage is mediated by helicase- and dsRNA termini-dependent states that are modulated by Loquacious-PD.Mol Cell. 2015 May 7;58(3):406-17. doi: 10.1016/j.molcel.2015.03.012. Epub 2015 Apr 16. Mol Cell. 2015. PMID: 25891075 Free PMC article.
-
Many ways to generate microRNA-like small RNAs: non-canonical pathways for microRNA production.Mol Genet Genomics. 2010 Aug;284(2):95-103. doi: 10.1007/s00438-010-0556-1. Epub 2010 Jul 2. Mol Genet Genomics. 2010. PMID: 20596726 Review.
-
Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.Nature. 2022 Jul;607(7918):399-406. doi: 10.1038/s41586-022-04911-x. Epub 2022 Jun 29. Nature. 2022. PMID: 35768513 Free PMC article.
-
Loqs-PD and R2D2 define independent pathways for RISC generation in Drosophila.Nucleic Acids Res. 2011 May;39(9):3836-51. doi: 10.1093/nar/gkq1324. Epub 2011 Jan 17. Nucleic Acids Res. 2011. PMID: 21245036 Free PMC article.
-
Specific requirement of DRB4, a dsRNA-binding protein, for the in vitro dsRNA-cleaving activity of Arabidopsis Dicer-like 4.RNA. 2011 Apr;17(4):750-60. doi: 10.1261/rna.2455411. Epub 2011 Jan 26. RNA. 2011. PMID: 21270136 Free PMC article.
References
-
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. - PubMed
-
- Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet. 2007;8:884–896. - PubMed
-
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
