Comparison of mitochondrial and nucleolar RNase MRP reveals identical RNA components with distinct enzymatic activities and protein components
- PMID: 20086051
- PMCID: PMC2822918
- DOI: 10.1261/rna.1893710
Comparison of mitochondrial and nucleolar RNase MRP reveals identical RNA components with distinct enzymatic activities and protein components
Abstract
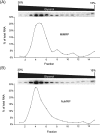
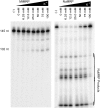
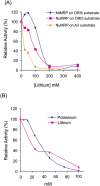
RNase MRP is a ribonucleoprotein endoribonuclease found in three cellular locations where distinct substrates are processed: the mitochondria, the nucleolus, and the cytoplasm. Cytoplasmic RNase MRP is the nucleolar enzyme that is transiently relocalized during mitosis. Nucleolar RNase MRP (NuMRP) was purified to homogeneity, and we extensively purified the mitochondrial RNase MRP (MtMRP) to a single RNA component identical to the NuMRP RNA. Although the protein components of the NuMRP were identified by mass spectrometry successfully, none of the known NuMRP proteins were found in the MtMRP preparation. Only trace amounts of the core NuMRP protein, Pop4, were detected in MtMRP by Western blot. In vitro activity of the two enzymes was compared. MtMRP cleaved only mitochondrial ORI5 substrate, while NuMRP cleaved all three substrates. However, the NuMRP enzyme cleaved the ORI5 substrate at sites different than the MtMRP enzyme. In addition, enzymatic differences in preferred ionic strength confirm these enzymes as distinct entities. Magnesium was found to be essential to both enzymes. We tested a number of reported inhibitors including puromycin, pentamidine, lithium, and pAp. Puromycin inhibition suggested that it binds directly to the MRP RNA, reaffirming the role of the RNA component in catalysis. In conclusion, our study confirms that the NuMRP and MtMRP enzymes are distinct entities with differing activities and protein components but a common RNA subunit, suggesting that the RNA must be playing a crucial role in catalytic activity.
Figures



Similar articles
-
Characterization of a unique protein component of yeast RNase MRP: an RNA-binding protein with a zinc-cluster domain.Genes Dev. 1994 Nov 1;8(21):2617-28. doi: 10.1101/gad.8.21.2617. Genes Dev. 1994. PMID: 7958920
-
Mutagenesis of SNM1, which encodes a protein component of the yeast RNase MRP, reveals a role for this ribonucleoprotein endoribonuclease in plasmid segregation.Mol Cell Biol. 1999 Nov;19(11):7857-69. doi: 10.1128/MCB.19.11.7857. Mol Cell Biol. 1999. PMID: 10523674 Free PMC article.
-
Basic domains target protein subunits of the RNase MRP complex to the nucleolus independently of complex association.Mol Biol Cell. 2001 Nov;12(11):3680-9. doi: 10.1091/mbc.12.11.3680. Mol Biol Cell. 2001. PMID: 11694598 Free PMC article.
-
The yeast, Saccharomyces cerevisiae, RNase P/MRP ribonucleoprotein endoribonuclease family.Mol Biol Rep. 1995-1996;22(2-3):87-93. doi: 10.1007/BF00988711. Mol Biol Rep. 1995. PMID: 8901493 Review.
-
Structural and functional similarities between MRP and RNase P.Mol Biol Rep. 1995-1996;22(2-3):81-5. doi: 10.1007/BF00988710. Mol Biol Rep. 1995. PMID: 8901492 Review.
Cited by
-
Non-coding RNA Regulated Cross-Talk Between Mitochondria and Other Cellular Compartments.Front Cell Dev Biol. 2021 Aug 3;9:688523. doi: 10.3389/fcell.2021.688523. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34414182 Free PMC article. Review.
-
PNPASE and RNA trafficking into mitochondria.Biochim Biophys Acta. 2012 Sep-Oct;1819(9-10):998-1007. doi: 10.1016/j.bbagrm.2011.10.001. Epub 2011 Oct 13. Biochim Biophys Acta. 2012. PMID: 22023881 Free PMC article. Review.
-
Substrate recognition by ribonucleoprotein ribonuclease MRP.RNA. 2011 Feb;17(2):356-64. doi: 10.1261/rna.2393711. Epub 2010 Dec 20. RNA. 2011. PMID: 21173200 Free PMC article.
-
Post-transcriptional diversity in riboproteins and RNAs in aging and cancer.Semin Cancer Biol. 2021 Nov;76:292-300. doi: 10.1016/j.semcancer.2021.08.012. Epub 2021 Aug 30. Semin Cancer Biol. 2021. PMID: 34474152 Free PMC article. Review.
-
The dichotomy of p53 regulation by noncoding RNAs.J Mol Cell Biol. 2014 Jun;6(3):198-205. doi: 10.1093/jmcb/mju017. Epub 2014 Apr 4. J Mol Cell Biol. 2014. PMID: 24706938 Free PMC article. Review.
References
-
- Boldogh IR, Pon LA. Purification and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 2007;80:45–64. - PubMed
-
- Brown RE, Jarvis KL, Hyland KJ. Protein measurement using bicinchoninic acid: Elimination of interfering substances. Anal Biochem. 1989;180:136–139. - PubMed
-
- Cai T, Schmitt ME. Characterization of ribonuclease MRP function. Methods Enzymol. 2001;342:135–142. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
