Yeast exonuclease 5 is essential for mitochondrial genome maintenance
- PMID: 20086101
- PMCID: PMC2832504
- DOI: 10.1128/MCB.01321-09
Yeast exonuclease 5 is essential for mitochondrial genome maintenance
Abstract
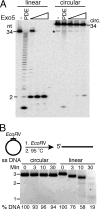
Yeast exonuclease 5 is encoded by the YBR163w (DEM1) gene, and this gene has been renamed EXO5. It is distantly related to the Escherichia coli RecB exonuclease class. Exo5 is localized to the mitochondria, and EXO5 deletions or nuclease-defective EXO5 mutants invariably yield petites, amplifying either the ori3 or ori5 region of the mitochondrial genome. These petites remain unstable and undergo continuous rearrangement. The mitochondrial phenotype of exo5Delta strains suggests an essential role for the enzyme in DNA replication and recombination. No nuclear phenotype associated with EXO5 deletions has been detected. Exo5 is a monomeric 5' exonuclease that releases dinucleotides as products. It is specific for single-stranded DNA and does not hydrolyze RNA. However, Exo5 has the capacity to slide across 5' double-stranded DNA or 5' RNA sequences and resumes cutting two nucleotides downstream of the double-stranded-to-single-stranded junction or RNA-to-DNA junction, respectively.
Figures





References
-
- Bernardi, G. 2005. Lessons from a small, dispensable genome: the mitochondrial genome of yeast. Gene 354:189-200. - PubMed
-
- Burgers, P. M. J., G. A. Bauer, and L. Tam. 1988. Exonuclease V from Saccharomyces cerevisiae. A 5′-3′-deoxyribonuclease that produces dinucleotides in a sequential fashion. J. Biol. Chem. 263:8099-8105. - PubMed
-
- Büttner, S., T. Eisenberg, D. Carmona-Gutierrez, D. Ruli, H. Knauer, C. Ruckenstuhl, C. Sigrist, S. Wissing, M. Kollroser, K. U. Frohlich, S. Sigrist, and F. Madeo. 2007. Endonuclease G regulates budding yeast life and death. Mol. Cell 25:233-246. - PubMed
-
- Bylund, G. O., J. Majka, and P. M. Burgers. 2006. Overproduction and purification of RFC-related clamp loaders and PCNA-related clamps from Saccharomyces cerevisiae. Methods Enzymol. 409:1-11. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
