Delineation of a bacterial starvation stress response network which can mediate antibiotic tolerance development
- PMID: 20086164
- PMCID: PMC2825962
- DOI: 10.1128/AAC.01218-09
Delineation of a bacterial starvation stress response network which can mediate antibiotic tolerance development
Abstract
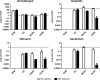
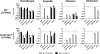
This study aimed at elucidating the physiological basis of bacterial antibiotic tolerance. By use of a combined phenotypic and gene knockout approach, exogenous nutrient composition was identified as a crucial environmental factor which could mediate progressive development of tolerance with markedly varied drug specificity and sustainability. Deprivation of amino acids was a prerequisite for tolerance formation, conferring condition-specific phenotypes against inhibitors of cell wall synthesis and DNA replication (ampicillin and ofloxacin, respectively), according to the relative abundances of ammonium salts, phosphate, and nucleobases. Upon further depletion of glucose, this variable phase consistently evolved into a sustainable mode, along with enhanced capacity to withstand the effect of the protein synthesis inhibitor gentamicin. Nevertheless, all phenotypes produced during spontaneous nutrient depletion lacked the sustainable, multidrug-tolerant features exhibited by the stationary-phase population and were attributed to complex interaction between starvation-mediated metabolic and stress protection responses on the basis of the following reasons: (i) the nutrition-dependent tolerance characteristics observed suggested that adaptive biosynthetic mechanisms could suppress but not fully avert tolerance under transient starvation conditions; (ii) formation of specific phenotypes could be inhibited by suppressing protein synthesis prior to nutrient depletion; (iii) bacteriostatic drugs produced only weak tolerance in the absence of starvation signals; and (iv) the attenuation of the stringent and SOS responses, as well as the functionality of other putative tolerance determinants, including rpoS, hipA, glpD, and phoU, could alter the induction requirement and drug specificity of the resultant phenotypes. These data reveal the common physiological grounds characteristic of starvation responses and the onset of antibiotic tolerance in bacteria.
Figures




Similar articles
-
PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli.Antimicrob Agents Chemother. 2007 Jun;51(6):2092-9. doi: 10.1128/AAC.00052-07. Epub 2007 Apr 9. Antimicrob Agents Chemother. 2007. PMID: 17420206 Free PMC article.
-
Starvation, together with the SOS response, mediates high biofilm-specific tolerance to the fluoroquinolone ofloxacin.PLoS Genet. 2013;9(1):e1003144. doi: 10.1371/journal.pgen.1003144. Epub 2013 Jan 3. PLoS Genet. 2013. PMID: 23300476 Free PMC article.
-
Transcription profiling of the stringent response in Escherichia coli.J Bacteriol. 2008 Feb;190(3):1084-96. doi: 10.1128/JB.01092-07. Epub 2007 Nov 26. J Bacteriol. 2008. PMID: 18039766 Free PMC article.
-
Kinase activity of overexpressed HipA is required for growth arrest and multidrug tolerance in Escherichia coli.J Bacteriol. 2006 Dec;188(24):8360-7. doi: 10.1128/JB.01237-06. Epub 2006 Oct 13. J Bacteriol. 2006. PMID: 17041039 Free PMC article.
-
Contribution of rpoS and bolA genes in biofilm formation in Escherichia coli K-12 MG1655.Mol Cell Biochem. 2010 Sep;342(1-2):207-13. doi: 10.1007/s11010-010-0485-7. Epub 2010 May 18. Mol Cell Biochem. 2010. PMID: 20480211 Review.
Cited by
-
Drug persistence - from antibiotics to cancer therapies.Curr Opin Syst Biol. 2018 Aug;10:1-8. doi: 10.1016/j.coisb.2018.03.003. Epub 2018 Mar 31. Curr Opin Syst Biol. 2018. PMID: 30740553 Free PMC article.
-
The role of metabolism in bacterial persistence.Front Microbiol. 2014 Mar 3;5:70. doi: 10.3389/fmicb.2014.00070. eCollection 2014. Front Microbiol. 2014. PMID: 24624123 Free PMC article. Review.
-
Active maintenance of proton motive force mediates starvation-induced bacterial antibiotic tolerance in Escherichia coli.Commun Biol. 2021 Sep 14;4(1):1068. doi: 10.1038/s42003-021-02612-1. Commun Biol. 2021. PMID: 34521984 Free PMC article.
-
Taking chances and making mistakes: non-genetic phenotypic heterogeneity and its consequences for surviving in dynamic environments.J R Soc Interface. 2017 Jul;14(132):20170141. doi: 10.1098/rsif.2017.0141. J R Soc Interface. 2017. PMID: 28701503 Free PMC article. Review.
-
Nitrogen Starvation Induces Persister Cell Formation in Escherichia coli.J Bacteriol. 2019 Jan 11;201(3):e00622-18. doi: 10.1128/JB.00622-18. Print 2019 Feb 1. J Bacteriol. 2019. PMID: 30420451 Free PMC article.
References
-
- Balaban, N. Q., J. Merrin, R. Chait, L. Kowalik, and S. Leibler. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622-1625. - PubMed
-
- Bigger, J. W. 1944. Treatment of staphylococcal infections with penicillin. Lancet ii:497-500.
-
- Debbia, E. A., S. Roveta, A. M. Schito, L. Gualco, and A. Marchese. 2001. Antibiotic persistence: the role of spontaneous DNA repair response. Microb. Drug Resist. 7:335-342. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

