Copy number abnormalities in sporadic canine colorectal cancers
- PMID: 20086242
- PMCID: PMC2840980
- DOI: 10.1101/gr.092726.109
Copy number abnormalities in sporadic canine colorectal cancers
Abstract
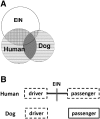
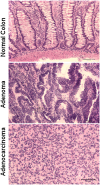
Human colorectal cancer (CRC) is one of the better-understood systems for studying the genetics of cancer initiation and progression. To develop a cross-species comparison strategy for identifying CRC causative gene or genomic alterations, we performed array comparative genomic hybridization (aCGH) to investigate copy number abnormalities (CNAs), one of the most prominent lesion types reported for human CRCs, in 10 spontaneously occurring canine CRCs. The results revealed for the first time a strong degree of genetic homology between sporadic canine and human CRCs. First, we saw that between 5% and 22% of the canine genome was amplified/deleted in these tumors, and that, reminiscent of human CRCs, the total altered sequences directly correlated to the tumor's progression stage, origin, and likely microsatellite instability status. Second, when mapping the identified CNAs onto syntenic regions of the human genome, we noted that the canine orthologs of genes participating in known human CRC pathways were recurrently disrupted, indicating that these pathways might be altered in the canine CRCs as well. Last, we observed a significant overlapping of CNAs between human and canine tumors, and tumors from the two species were clustered according to the tumor subtypes but not the species. Significantly, compared with the shared CNAs, we found that species-specific (especially human-specific) CNAs localize to evolutionarily unstable regions that harbor more segmental duplications and interspecies genomic rearrangement breakpoints. These findings indicate that CNAs recurrent between human and dog CRCs may have a higher probability of being cancer-causative, compared with CNAs found in one species only.
Figures

 , where l and m are the total probe number and the mean log2 ratio of the CNA. Except for CNAs that are larger than 1 Mb in size, the width of the vertical lines is not drawn to scale with the chromosome length. (B) Mapping dog CNAs onto the human genome. A total of 9107 CNAs and 541.6 Mb (98.2% of the total) of the same dog tumor shown above were mapped onto the human genome, amounting to 609 Mb on the human genome.
, where l and m are the total probe number and the mean log2 ratio of the CNA. Except for CNAs that are larger than 1 Mb in size, the width of the vertical lines is not drawn to scale with the chromosome length. (B) Mapping dog CNAs onto the human genome. A total of 9107 CNAs and 541.6 Mb (98.2% of the total) of the same dog tumor shown above were mapped onto the human genome, amounting to 609 Mb on the human genome.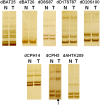

 , where dij is the distance between a tumor Ti of cluster X and a tumor Tj of cluster Y calculated as described in the text, and |X| and |Y| are the total number of tumors inside clusters X and Y, respectively. (B) Clustering of tumors from both humans and dogs. The tree was constructed as described above, using the overlapping information of CNAs either identified on (for the human tumors T2551 and T3912) or mapped onto (for the dog tumors, see Fig. 3) the human genome. MSI-L: MSI-low; MSI-H: MSI-high.
, where dij is the distance between a tumor Ti of cluster X and a tumor Tj of cluster Y calculated as described in the text, and |X| and |Y| are the total number of tumors inside clusters X and Y, respectively. (B) Clustering of tumors from both humans and dogs. The tree was constructed as described above, using the overlapping information of CNAs either identified on (for the human tumors T2551 and T3912) or mapped onto (for the dog tumors, see Fig. 3) the human genome. MSI-L: MSI-low; MSI-H: MSI-high.Similar articles
-
Functionally-focused algorithmic analysis of high resolution microarray-CGH genomic landscapes demonstrates comparable genomic copy number aberrations in MSI and MSS sporadic colorectal cancer.PLoS One. 2017 Feb 23;12(2):e0171690. doi: 10.1371/journal.pone.0171690. eCollection 2017. PLoS One. 2017. PMID: 28231327 Free PMC article.
-
Association between genomic alterations and metastatic behavior of colorectal cancer identified by array-based comparative genomic hybridization.Genes Chromosomes Cancer. 2013 Feb;52(2):140-9. doi: 10.1002/gcc.22013. Epub 2012 Oct 17. Genes Chromosomes Cancer. 2013. PMID: 23073979
-
Array CGH identifies distinct DNA copy number profiles of oncogenes and tumor suppressor genes in chromosomal- and microsatellite-unstable sporadic colorectal carcinomas.J Mol Med (Berl). 2007 Mar;85(3):293-304. doi: 10.1007/s00109-006-0126-5. Epub 2006 Dec 2. J Mol Med (Berl). 2007. PMID: 17143621
-
Genetics and Genetic Biomarkers in Sporadic Colorectal Cancer.Gastroenterology. 2015 Oct;149(5):1177-1190.e3. doi: 10.1053/j.gastro.2015.06.047. Epub 2015 Jul 26. Gastroenterology. 2015. PMID: 26216840 Free PMC article. Review.
-
Somatic gene copy number alterations in colorectal cancer: new quest for cancer drivers and biomarkers.Oncogene. 2016 Apr 21;35(16):2011-9. doi: 10.1038/onc.2015.304. Epub 2015 Aug 10. Oncogene. 2016. PMID: 26257062 Review.
Cited by
-
Identification of RECK as an evolutionarily conserved tumor suppressor gene for zebrafish malignant peripheral nerve sheath tumors.Oncotarget. 2018 May 4;9(34):23494-23504. doi: 10.18632/oncotarget.25236. eCollection 2018 May 4. Oncotarget. 2018. PMID: 29805750 Free PMC article.
-
Origins of the domestic dog and the rich potential for gene mapping.Genet Res Int. 2011;2011:579308. doi: 10.4061/2011/579308. Epub 2011 Jan 17. Genet Res Int. 2011. PMID: 22567358 Free PMC article.
-
Intestinal Stem Cells to Advance Drug Development, Precision, and Regenerative Medicine: A Paradigm Shift in Translational Research.AAPS J. 2017 Dec 12;20(1):17. doi: 10.1208/s12248-017-0178-1. AAPS J. 2017. PMID: 29234895 Free PMC article. Review.
-
Immunohistochemical expression of β-catenin, Ki67, CD3 and CD18 in canine colorectal adenomas and adenocarcinomas.BMC Vet Res. 2021 Mar 12;17(1):119. doi: 10.1186/s12917-021-02829-6. BMC Vet Res. 2021. PMID: 33712002 Free PMC article.
-
Molecular homology and difference between spontaneous canine mammary cancer and human breast cancer.Cancer Res. 2014 Sep 15;74(18):5045-56. doi: 10.1158/0008-5472.CAN-14-0392. Epub 2014 Jul 31. Cancer Res. 2014. PMID: 25082814 Free PMC article.
References
-
- Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–376. - PubMed
-
- Argyle DJ. The benefits of comparative medicine—a hundred years to come to our senses. Vet J. 2005;170:147–148. - PubMed
-
- Argyle DJ. Prostate cancer in dogs and men: A unique opportunity to study the disease. Vet J. 2009;180:137–138. - PubMed
-
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. - PubMed
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
