The full-length Saccharomyces cerevisiae Sgs1 protein is a vigorous DNA helicase that preferentially unwinds holliday junctions
- PMID: 20086270
- PMCID: PMC2832980
- DOI: 10.1074/jbc.M109.083196
The full-length Saccharomyces cerevisiae Sgs1 protein is a vigorous DNA helicase that preferentially unwinds holliday junctions
Abstract
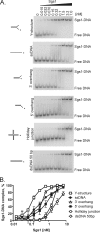
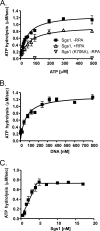
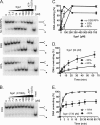
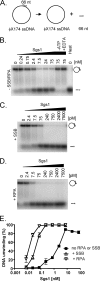
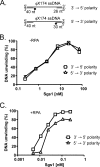
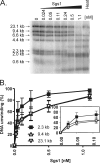
The highly conserved RecQ family of DNA helicases has multiple roles in the maintenance of genome stability. Sgs1, the single RecQ homologue in Saccharomyces cerevisiae, acts both early and late during homologous recombination. Here we present the expression, purification, and biochemical analysis of full-length Sgs1. Unlike the truncated form of Sgs1 characterized previously, full-length Sgs1 binds diverse single-stranded and double-stranded DNA substrates, including DNA duplexes with 5'- and 3'-single-stranded DNA overhangs. Similarly, Sgs1 unwinds a variety of DNA substrates, including blunt-ended duplex DNA. Significantly, a substrate containing a Holliday junction is unwound most efficiently. DNA unwinding is catalytic, requires ATP, and is stimulated by replication protein A. Unlike RecQ homologues from multicellular organisms, Sgs1 is remarkably active at picomolar concentrations and can efficiently unwind duplex DNA molecules as long as 23,000 base pairs. Our analysis shows that Sgs1 resembles Escherichia coli RecQ protein more than any of the human RecQ homologues with regard to its helicase activity. The full-length recombinant protein will be invaluable for further investigation of Sgs1 biochemistry.
Figures


References
-
- Nakayama H., Nakayama K., Nakayama R., Irino N., Nakayama Y., Hanawalt P. C. (1984) Mol. Gen. Genet. 195, 474–480 - PubMed
-
- Watt P. M., Louis E. J., Borts R. H., Hickson I. D. (1995) Cell 81, 253–260 - PubMed
-
- Wu L., Hickson I. D. (2006) Annu. Rev. Genet. 40, 279–306 - PubMed
-
- Ellis N. A., Groden J., Ye T. Z., Straughen J., Lennon D. J., Ciocci S., Proytcheva M., German J. (1995) Cell 83, 655–666 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

