Large meta-analysis of multiple cancers reveals a common, compact and highly prognostic hypoxia metagene
- PMID: 20087356
- PMCID: PMC2816644
- DOI: 10.1038/sj.bjc.6605450
Large meta-analysis of multiple cancers reveals a common, compact and highly prognostic hypoxia metagene
Erratum in
- Br J Cancer. 2010 Sep 7;103(6):929
Abstract
Background: There is a need to develop robust and clinically applicable gene expression signatures. Hypoxia is a key factor promoting solid tumour progression and resistance to therapy; a hypoxia signature has the potential to be not only prognostic but also to predict benefit from particular interventions.
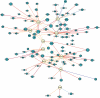
Methods: An approach for deriving signatures that combine knowledge of gene function and analysis of in vivo co-expression patterns was used to define a common hypoxia signature from three head and neck and five breast cancer studies. Previously validated hypoxia-regulated genes (seeds) were used to generate hypoxia co-expression cancer networks.
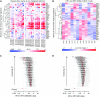
Results: A common hypoxia signature, or metagene, was derived by selecting genes that were consistently co-expressed with the hypoxia seeds in multiple cancers. This was highly enriched for hypoxia-regulated pathways, and prognostic in multivariate analyses. Genes with the highest connectivity were also the most prognostic, and a reduced metagene consisting of a small number of top-ranked genes, including VEGFA, SLC2A1 and PGAM1, outperformed both a larger signature and reported signatures in independent data sets of head and neck, breast and lung cancers.
Conclusion: Combined knowledge of multiple genes' function from in vitro experiments together with meta-analysis of multiple cancers can deliver compact and robust signatures suitable for clinical application.
Figures

Comment in
-
Cancer: Suffocation of gene expression.Nature. 2016 Sep 1;537(7618):42-43. doi: 10.1038/nature19426. Epub 2016 Aug 17. Nature. 2016. PMID: 27533033 No abstract available.
References
-
- Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, Lizyness ML, Kuick R, Hayasaka S, Taylor JM, Iannettoni MD, Orringer MB, Hanash S (2002) Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med 8: 816–824 - PubMed
-
- Butte AJ, Kohane IS (2003) Relevance networks: a first step towards finding genetic regulatory networks within microarray data. In The Analysis of Gene Expression Data Parmigiani G, Gar-rett ES, Irizarry RA, Zeger S (eds). Springer-Verlag: New York
-
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M (2006) Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38: 1289–1297 - PubMed
-
- Chi JT, Wang Z, Nuyten DS, Rodriguez EH, Schaner ME, Salim A, Wang Y, Kristensen GB, Helland A, Børresen-Dale AL, Giaccia A, Longaker MT, Hastie T, Yang GP, van de Vijver MJ, Brown PO (2006) Gene expression programs in response to hypoxia: cell type specificity and prognostic significance in human cancers. PLoS Med 3: e47. - PMC - PubMed
-
- Choi P, Chen C (2005) Genetic expression profiles and biologic pathway alterations in head and neck squamous cell carcinoma. Cancer 104: 1113–1128 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

