Crystallographic studies of chemically modified nucleic acids: a backward glance
- PMID: 20087997
- PMCID: PMC2905155
- DOI: 10.1002/cbdv.200900177
Crystallographic studies of chemically modified nucleic acids: a backward glance
Abstract
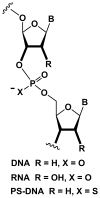
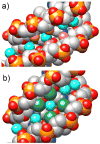
Chemically modified nucleic acids (CNAs) are widely explored as antisense oligonucleotide or small interfering RNA (siRNA) candidates for therapeutic applications. CNAs are also of interest in diagnostics, high-throughput genomics and target validation, nanotechnology and as model systems in investigations directed at a better understanding of the etiology of nucleic acid structure, as well as the physicochemical and pairing properties of DNA and RNA, and for probing protein-nucleic acid interactions. In this article, we review research conducted in our laboratory over the past two decades with a focus on crystal-structure analyses of CNAs and artificial pairing systems. We highlight key insights into issues ranging from conformational distortions as a consequence of modification to the modulation of pairing strength, and RNA affinity by stereoelectronic effects and hydration. Although crystal structures have only been determined for a subset of the large number of modifications that were synthesized and analyzed in the oligonucleotide context to date, they have yielded guiding principles for the design of new analogs with tailor-made properties, including pairing specificity, nuclease resistance, and cellular uptake. And, perhaps less obviously, crystallographic studies of CNAs and synthetic pairing systems have shed light on fundamental aspects of DNA and RNA structure and function that would not have been disclosed by investigations solely focused on the natural nucleic acids.
Figures
Similar articles
-
Insights from crystallographic studies into the structural and pairing properties of nucleic acid analogs and chemically modified DNA and RNA oligonucleotides.Annu Rev Biophys Biomol Struct. 2007;36:281-305. doi: 10.1146/annurev.biophys.36.040306.132556. Annu Rev Biophys Biomol Struct. 2007. PMID: 17288535 Review.
-
Re-Engineering RNA Molecules into Therapeutic Agents.Acc Chem Res. 2019 Apr 16;52(4):1036-1047. doi: 10.1021/acs.accounts.8b00650. Epub 2019 Mar 26. Acc Chem Res. 2019. PMID: 30912917
-
Probing the Backbone Topology of DNA: Synthesis and Properties of 7',5'-Bicyclo-DNA.Chemistry. 2017 Jun 12;23(33):7953-7968. doi: 10.1002/chem.201700435. Epub 2017 Apr 10. Chemistry. 2017. PMID: 28262999
-
Comparison of duplex stabilizing properties of 2'-fluorinated nucleic acid analogues with furanose and non-furanose sugar rings.J Org Chem. 2014 Sep 19;79(18):8877-81. doi: 10.1021/jo501381q. Epub 2014 Aug 27. J Org Chem. 2014. PMID: 25137618
-
Exploring the role of chirality in nucleic acid recognition.Chem Biodivers. 2011 Mar;8(3):373-413. doi: 10.1002/cbdv.201000303. Chem Biodivers. 2011. PMID: 21404424 Review.
Cited by
-
Structural insight into DNA-assembled oligochromophores: crystallographic analysis of pyrene- and phenanthrene-modified DNA in complex with BpuJI endonuclease.Nucleic Acids Res. 2016 Sep 6;44(15):7079-89. doi: 10.1093/nar/gkw644. Epub 2016 Jul 15. Nucleic Acids Res. 2016. PMID: 27422870 Free PMC article.
-
Same fold, different properties: polarizable molecular dynamics simulations of telomeric and TERRA G-quadruplexes.Nucleic Acids Res. 2020 Jan 24;48(2):561-575. doi: 10.1093/nar/gkz1154. Nucleic Acids Res. 2020. PMID: 31807754 Free PMC article.
-
NMR solution structure of tricyclo-DNA containing duplexes: insight into enhanced thermal stability and nuclease resistance.Nucleic Acids Res. 2019 May 21;47(9):4872-4882. doi: 10.1093/nar/gkz197. Nucleic Acids Res. 2019. PMID: 30916334 Free PMC article.
-
Tellurium-Modified Nucleosides, Nucleotides, and Nucleic Acids with Potential Applications.Molecules. 2022 Dec 1;27(23):8379. doi: 10.3390/molecules27238379. Molecules. 2022. PMID: 36500495 Free PMC article. Review.
-
The many twists and turns of DNA: template, telomere, tool, and target.Curr Opin Struct Biol. 2010 Jun;20(3):262-75. doi: 10.1016/j.sbi.2010.03.001. Epub 2010 Apr 8. Curr Opin Struct Biol. 2010. PMID: 20381338 Free PMC article. Review.
References
-
- Uhlmann E, Peyman A. Chem Rev. 1990;90:543.
-
- Milligan JF, Matteucci MD, Martin JC. J Med Chem. 1993;36:1923. - PubMed
-
- De Mesmaeker A, Häner R, Martin P, Moser HE. Acc Chem Res. 1995;28:366.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

