The hunt for 8-oxoguanine deaminase
- PMID: 20088583
- PMCID: PMC2820149
- DOI: 10.1021/ja909817d
The hunt for 8-oxoguanine deaminase
Abstract
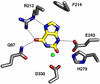
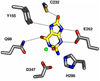
An enzyme from Pseudomonas aeruginosa, Pa0142 (gi|9945972), that is able to catalyze the deamination of 8-oxoguanine (8-oxoG) to uric acid has been identified for the first time. 8-Oxoguanine is formed by the oxidation of guanine residues within DNA by reactive oxygen species, and this lesion results in G:C to T:A transversions. The value of k(cat)/K(m) for the deamination of 8-oxoG by Pa0142 at pH 8.0 and 30 degrees C is 2.0 x 10(4) M(-1) s(-1). This enzyme can also catalyze the deamination of isocystosine and guanine at rates that are approximately an order of magnitude lower. The three-dimensional structure of a homologous enzyme (gi|44264246) from the Sargasso Sea has been determined by X-ray diffraction methods to a resolution of 2.2 A (PDB entry). The enzyme folds as a (beta/alpha)(8) barrel and is a member of the amidohydrolase superfamily with a single zinc in the active site. This enzyme catalyzes the deamination of 8-oxoG with a k(cat)/K(m) value of 2.7 x 10(5) M(-1) s(-1). Computational docking of potential high-energy intermediates for the deamination reaction to the X-ray crystal structure suggests that active-site binding of 8-oxoG is facilitated by hydrogen-bond interactions from a conserved glutamine that follows beta-strand 1 with the carbonyl group at C6, a conserved tyrosine that follows beta-strand 2 with N7, and a conserved cysteine residue that follows beta-strand 4 with the carbonyl group at C8. A bioinformatic analysis of available protein sequences suggests that approximately 200 other bacteria possess an enzyme capable of catalyzing the deamination of 8-oxoG.
Figures

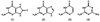
Similar articles
-
Rescue of the orphan enzyme isoguanine deaminase.Biochemistry. 2011 Jun 28;50(25):5555-7. doi: 10.1021/bi200680y. Epub 2011 Jun 7. Biochemistry. 2011. PMID: 21604715 Free PMC article.
-
Discovery and structure determination of the orphan enzyme isoxanthopterin deaminase.Biochemistry. 2010 May 25;49(20):4374-82. doi: 10.1021/bi100252s. Biochemistry. 2010. PMID: 20415463 Free PMC article.
-
Bacterial ammeline metabolism via guanine deaminase.J Bacteriol. 2010 Feb;192(4):1106-12. doi: 10.1128/JB.01243-09. Epub 2009 Dec 18. J Bacteriol. 2010. PMID: 20023034 Free PMC article.
-
Multiple DNA glycosylases for repair of 8-oxoguanine and their potential in vivo functions.Prog Nucleic Acid Res Mol Biol. 2001;68:193-205. doi: 10.1016/s0079-6603(01)68100-5. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11554297 Review.
-
8-oxoguanine DNA glycosylases: one lesion, three subfamilies.Int J Mol Sci. 2012;13(6):6711-6729. doi: 10.3390/ijms13066711. Epub 2012 Jun 1. Int J Mol Sci. 2012. PMID: 22837659 Free PMC article. Review.
Cited by
-
Discovery of a bacterial 5-methylcytosine deaminase.Biochemistry. 2014 Dec 2;53(47):7426-35. doi: 10.1021/bi5012767. Epub 2014 Nov 19. Biochemistry. 2014. PMID: 25384249 Free PMC article.
-
Rescue of the orphan enzyme isoguanine deaminase.Biochemistry. 2011 Jun 28;50(25):5555-7. doi: 10.1021/bi200680y. Epub 2011 Jun 7. Biochemistry. 2011. PMID: 21604715 Free PMC article.
-
The structure of the hexameric atrazine chlorohydrolase AtzA.Acta Crystallogr D Biol Crystallogr. 2015 Mar;71(Pt 3):710-20. doi: 10.1107/S1399004715000619. Epub 2015 Feb 26. Acta Crystallogr D Biol Crystallogr. 2015. PMID: 25760618 Free PMC article.
-
Nucleobase deaminases: a potential enzyme system for new therapies.RSC Adv. 2018 Jun 28;8(42):23567-23577. doi: 10.1039/c8ra04112a. eCollection 2018 Jun 27. RSC Adv. 2018. PMID: 35540270 Free PMC article. Review.
-
Identification of a 2'-O-Methyluridine Nucleoside Hydrolase Using the Metagenomic Libraries.Molecules. 2018 Nov 7;23(11):2904. doi: 10.3390/molecules23112904. Molecules. 2018. PMID: 30405065 Free PMC article.
References
-
- Seibert CM, Raushel FM. Biochemistry. 2005;44:6383–6391. - PubMed
-
- Structural Genomics Consortium. 2007 (pdb code: 2uz9)
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

