Analysis of monomeric and dimeric phosphorylated forms of protein kinase R
- PMID: 20088595
- PMCID: PMC2819637
- DOI: 10.1021/bi901873p
Analysis of monomeric and dimeric phosphorylated forms of protein kinase R
Abstract
PKR (protein kinase R) is induced by interferon and is a key component of the innate immunity antiviral pathway. Upon binding double-stranded RNA (dsRNA) or dimerization in the absence of dsRNA, PKR undergoes autophosphorylation at multiple serines and threonines that activate the kinase. Although it has previously been demonstrated that phosphorylation enhances PKR dimerization, gel filtration analysis reveals a second monomeric phosphorylated form. These forms are termed phosphorylated dimeric PKR (pPKRd) and phosphorylated monomeric PKR (pPKRm). These two forms do not reversibly interconvert. Sedimentation equilibrium measurements reveal that pPKRm dimerizes weakly with a K(d) similar to that of unphosphorylated PKR. Isoelectric focusing and mass spectrometry demonstrate that both pPKRm and pPKRd are heterogeneous in their phosphorylation states, with an average of 9 or 10 phosphates. Equilibrium chemical denaturation analysis indicates that phosphorylation destabilizes the kinase domain by approximately 1.5 kcal/mol in the dimeric form but not in the monomeric form. Limited proteolysis also reveals that phosphorylation induces a conformational change in pPKRd that is not detected in pPKRm. pPKRm binds dsRNA with an affinity similar to that of unphosphorylated PKR, whereas binding cannot be detected with pPKRd. Despite these substantial differences in biophysical properties, both pPKRm and pPKRd are catalytically competent and are activated to phosphorylate the PKR substrate eIF2alpha in the absence of dsRNA. Thus, both monomeric and dimeric forms of phosphorylated PKR may participate in the interferon antiviral pathway.
Figures
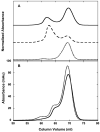

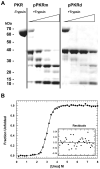
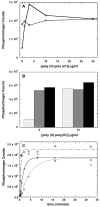
 ) and pPKRm (
) and pPKRm ( ). B) Comparison of unphosphorylated PKR (white), pPKRm (grey) and pPKRd (black). For both A and B, samples contained 400 μM ATP and were quenched after 20 minutes. C) Kinetics of eIF2α phosphorylation catalyzed by unphosphorylated PKR (
). B) Comparison of unphosphorylated PKR (white), pPKRm (grey) and pPKRd (black). For both A and B, samples contained 400 μM ATP and were quenched after 20 minutes. C) Kinetics of eIF2α phosphorylation catalyzed by unphosphorylated PKR ( ), pPKRm (
), pPKRm ( ) and pPKRd (
) and pPKRd ( ). All samples contained 40 μM ATP and the unphosphorylated PKR sample contained 10 μg/ml dsRNA. The best fit line for a single exponential fit is shown for each reaction, with rate constants of 0.64 min−1 (PKR), 0.22 min−1 (pPKRm) and 2.2 min−1 (pPKRd).
). All samples contained 40 μM ATP and the unphosphorylated PKR sample contained 10 μg/ml dsRNA. The best fit line for a single exponential fit is shown for each reaction, with rate constants of 0.64 min−1 (PKR), 0.22 min−1 (pPKRm) and 2.2 min−1 (pPKRd).Similar articles
-
Mechanism of PKR activation: dimerization and kinase activation in the absence of double-stranded RNA.J Mol Biol. 2005 Jan 7;345(1):81-90. doi: 10.1016/j.jmb.2004.10.031. J Mol Biol. 2005. PMID: 15567412
-
Mechanistic link between PKR dimerization, autophosphorylation, and eIF2alpha substrate recognition.Cell. 2005 Sep 23;122(6):901-13. doi: 10.1016/j.cell.2005.06.041. Cell. 2005. PMID: 16179259
-
Homologous regions of the alpha subunit of eukaryotic translational initiation factor 2 (eIF2alpha) and the vaccinia virus K3L gene product interact with the same domain within the dsRNA-activated protein kinase (PKR).Eur J Biochem. 1997 Nov 15;250(1):85-91. doi: 10.1111/j.1432-1033.1997.00085.x. Eur J Biochem. 1997. PMID: 9431994
-
PKR and eIF2alpha: integration of kinase dimerization, activation, and substrate docking.Cell. 2005 Sep 23;122(6):823-5. doi: 10.1016/j.cell.2005.09.007. Cell. 2005. PMID: 16179248 Review.
-
Protein kinase PKR and RNA adenosine deaminase ADAR1: new roles for old players as modulators of the interferon response.Curr Opin Immunol. 2011 Oct;23(5):573-82. doi: 10.1016/j.coi.2011.08.009. Epub 2011 Sep 15. Curr Opin Immunol. 2011. PMID: 21924887 Free PMC article. Review.
Cited by
-
Heparin activates PKR by inducing dimerization.J Mol Biol. 2011 Nov 11;413(5):973-84. doi: 10.1016/j.jmb.2011.09.025. Epub 2011 Sep 28. J Mol Biol. 2011. PMID: 21978664 Free PMC article.
-
Analysis of PKR activation using analytical ultracentrifugation.Macromol Biosci. 2010 Jul 7;10(7):703-13. doi: 10.1002/mabi.201000069. Macromol Biosci. 2010. PMID: 20533534 Free PMC article.
-
Raf kinase inhibitor protein (RKIP) dimer formation controls its target switch from Raf1 to G protein-coupled receptor kinase (GRK) 2.J Biol Chem. 2012 Jul 6;287(28):23407-17. doi: 10.1074/jbc.M112.363812. Epub 2012 May 17. J Biol Chem. 2012. PMID: 22610096 Free PMC article.
-
Mechanism of Protein Kinase R Inhibition by Human Cytomegalovirus pTRS1.J Virol. 2017 Feb 14;91(5):e01574-16. doi: 10.1128/JVI.01574-16. Print 2017 Mar 1. J Virol. 2017. PMID: 27974558 Free PMC article.
References
-
- Kaufman RJ. The double stranded RNA-activated protein kinase PKR. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2000. pp. 503–528.
-
- Toth AM, Zhang P, Das S, George CX, Samuel CE. Interferon action and the double-stranded RNA-dependent enzymes ADAR1 adenosine deaminase and PKR protein kinase. Prog. Nucleic Acid Res. Mol. Biol. 2006;81:369–434. - PubMed
-
- Koromilas AE, Roy S, Barber GN, Katze MG, Sonenberg N. Malignant transformation by a mutant of the IFN-inducible dsRNA-dependent protein kinase. Science. 1992;257:1685–1689. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

