Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus
- PMID: 20088951
- PMCID: PMC7159092
- DOI: 10.1111/j.1462-5822.2010.01437.x
Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus
Abstract
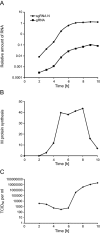
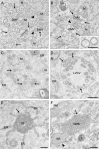
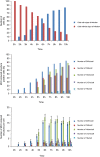
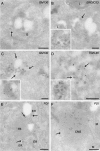
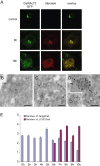
Coronaviruses (CoV) are enveloped positive-strand RNA viruses that induce different membrane rearrangements in infected cells in order to efficiently replicate and assemble. The origin, the protein composition and the function of these structures are not well established. To shed further light on these structures, we have performed a time-course experiment in which the mouse hepatitis virus (MHV)-induced membrane rearrangements were examined qualitatively and quantitatively by (immuno)-electron microscopy. With our approach we were able to confirm the appearance of 6, previously reported, membranous structures during the course of a complete infection cycle. These structures include the well-characterized double-membrane vesicles (DMVs), convoluted membranes (CMs) and virions but also the more enigmatic large virion-containing vacuoles (LVCVs), tubular bodies (TBs) and cubic membrane structures (CMSs). We have characterized the LVCVs, TBs and CMSs, and found that the CoV-induced structures appear in a strict order. By combining these data with quantitative analyses on viral RNA, protein synthesis and virion release, this study generates an integrated molecular and ultrastructural overview of CoV infection. In particular, it provides insights in the role of each CoV-induced structure and reveals that LVCVs are ERGIC/Golgi compartments that expand to accommodate an increasing production of viral particles.
Figures


References
-
- Almsherqi, Z.A. , McLachlan, C.S. , Mossop, P. , Knoops, K. , and Deng, Y. (2005) Direct template matching reveals a host subcellular membrane gyroid cubic structure that is associated with SARS virus. Redox Rep 10: 167–171. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

