Genetic relationships among seven sections of genus Arachis studied by using SSR markers
- PMID: 20089171
- PMCID: PMC2826335
- DOI: 10.1186/1471-2229-10-15
Genetic relationships among seven sections of genus Arachis studied by using SSR markers
Abstract
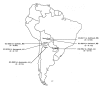
Background: The genus Arachis, originated in South America, is divided into nine taxonomical sections comprising of 80 species. Most of the Arachis species are diploids (2n = 2x = 20) and the tetraploid species (2n = 2x = 40) are found in sections Arachis, Extranervosae and Rhizomatosae. Diploid species have great potential to be used as resistance sources for agronomic traits like pests and diseases, drought related traits and different life cycle spans. Understanding of genetic relationships among wild species and between wild and cultivated species will be useful for enhanced utilization of wild species in improving cultivated germplasm. The present study was undertaken to evaluate genetic relationships among species (96 accessions) belonging to seven sections of Arachis by using simple sequence repeat (SSR) markers developed from Arachis hypogaea genomic library and gene sequences from related genera of Arachis.
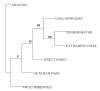
Results: The average transferability rate of 101 SSR markers tested to section Arachis and six other sections was 81% and 59% respectively. Five markers (IPAHM 164, IPAHM 165, IPAHM 407a, IPAHM 409, and IPAHM 659) showed 100% transferability. Cluster analysis of allelic data from a subset of 32 SSR markers on 85 wild and 11 cultivated accessions grouped accessions according to their genome composition, sections and species to which they belong. A total of 109 species specific alleles were detected in different wild species, Arachis pusilla exhibited largest number of species specific alleles (15). Based on genetic distance analysis, the A-genome accession ICG 8200 (A. duranensis) and the B-genome accession ICG 8206 (A. ipaënsis) were found most closely related to A. hypogaea.
Conclusion: A set of cross species and cross section transferable SSR markers has been identified that will be useful for genetic studies of wild species of Arachis, including comparative genome mapping, germplasm analysis, population genetic structure and phylogenetic inferences among species. The present study provides strong support based on both genomic and genic markers, probably for the first time, on relationships of A. monticola and A. hypogaea as well as on the most probable donor of A and B-genomes of cultivated groundnut.
Figures

Similar articles
-
Genetic diversity of peanut (Arachis hypogaea L.) and its wild relatives based on the analysis of hypervariable regions of the genome.BMC Plant Biol. 2004 Jul 14;4:11. doi: 10.1186/1471-2229-4-11. BMC Plant Biol. 2004. PMID: 15253775 Free PMC article.
-
Comparative mapping in intraspecific populations uncovers a high degree of macrosynteny between A- and B-genome diploid species of peanut.BMC Genomics. 2012 Nov 10;13:608. doi: 10.1186/1471-2164-13-608. BMC Genomics. 2012. PMID: 23140574 Free PMC article.
-
RAPD and ISSR fingerprints as useful genetic markers for analysis of genetic diversity, varietal identification, and phylogenetic relationships in peanut (Arachis hypogaea) cultivars and wild species.Genome. 2001 Oct;44(5):763-72. Genome. 2001. PMID: 11681599
-
A study of the relationships of cultivated peanut (Arachis hypogaea) and its most closely related wild species using intron sequences and microsatellite markers.Ann Bot. 2013 Jan;111(1):113-26. doi: 10.1093/aob/mcs237. Epub 2012 Nov 6. Ann Bot. 2013. PMID: 23131301 Free PMC article.
-
Abundant microsatellite diversity and oil content in wild Arachis species.PLoS One. 2012;7(11):e50002. doi: 10.1371/journal.pone.0050002. Epub 2012 Nov 20. PLoS One. 2012. PMID: 23185514 Free PMC article.
Cited by
-
Genetic imprints of domestication for disease resistance, oil quality, and yield component traits in groundnut (Arachis hypogaea L.).Mol Genet Genomics. 2019 Apr;294(2):365-378. doi: 10.1007/s00438-018-1511-9. Epub 2018 Nov 22. Mol Genet Genomics. 2019. PMID: 30467595
-
Development of an oligonucleotide dye solution facilitates high throughput and cost-efficient chromosome identification in peanut.Plant Methods. 2019 Jul 8;15:69. doi: 10.1186/s13007-019-0451-7. eCollection 2019. Plant Methods. 2019. PMID: 31316581 Free PMC article.
-
Genome-Wide Discovery and Deployment of Insertions and Deletions Markers Provided Greater Insights on Species, Genomes, and Sections Relationships in the Genus Arachis.Front Plant Sci. 2017 Dec 19;8:2064. doi: 10.3389/fpls.2017.02064. eCollection 2017. Front Plant Sci. 2017. PMID: 29312366 Free PMC article.
-
Chloroplast Phylogenomic Analyses Reveal a Maternal Hybridization Event Leading to the Formation of Cultivated Peanuts.Front Plant Sci. 2021 Dec 17;12:804568. doi: 10.3389/fpls.2021.804568. eCollection 2021. Front Plant Sci. 2021. PMID: 34975994 Free PMC article.
-
Marker-assisted introgression of wild chromosome segments conferring resistance to fungal foliar diseases into peanut (Arachis hypogaea L.).Front Plant Sci. 2023 Mar 17;14:1139361. doi: 10.3389/fpls.2023.1139361. eCollection 2023. Front Plant Sci. 2023. PMID: 37056498 Free PMC article.
References
-
- Krapovickas A, Gregory WC. Taxonomia del género Arachis (Leguminosae) Bonplandia. 1994;8:1–186.
-
- Valls JFM, Simpson CE. New species of Arachis from Brazil, Paraguay, and Bolivia. Bonplandia. 2005;14:35–64.
-
- Murty UR, Jahnavi MR, Bharati M, Kirti PB. In: Proceedings of International Workshop on Cytogenetics of Arachis: 31 Oct-2 Nov 1983. Moss JP, editor. ICRISAT; 1985. Chromosome morphology and Gregory's sectional delimitation in the genus Arachis L; pp. 81–83.
-
- Jahnavi MR, Murty UR. Chromosome morphology in species of the sections Erectoides and Extranervosae of the genus Arachis L. Cytologia. 1985;50:747–758.
-
- Valls JFM, Simpson CE. In: Biology and agronomy of forage Arachis. Kerridge PC, Hardy B, editor. Cali, CIAT; 1994. Taxonomy, natural distribution, and attributes of Arachis; pp. 1–18.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

