A milieu of regulatory elements in the epidermal differentiation complex syntenic block: implications for atopic dermatitis and psoriasis
- PMID: 20089530
- PMCID: PMC2846160
- DOI: 10.1093/hmg/ddq019
A milieu of regulatory elements in the epidermal differentiation complex syntenic block: implications for atopic dermatitis and psoriasis
Abstract
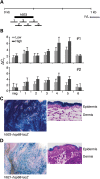
Two common inflammatory skin disorders with impaired barrier, atopic dermatitis (AD) and psoriasis, share distinct genetic linkage to the Epidermal Differentiation Complex (EDC) locus on 1q21. The EDC is comprised of tandemly arrayed gene families encoding proteins involved in skin cell differentiation. Discovery of semi-dominant mutations in filaggrin (FLG) associated with AD and a copy number variation within the LCE genes associated with psoriasis provide compelling evidence for the role of EDC genes in the pathogenesis of these diseases. To date, little is known about the potentially complex regulatory landscape within the EDC. Here, we report a computational approach to identify conserved non-coding elements (CNEs) in the EDC queried for regulatory function. Coordinate expression of EDC genes during mouse embryonic skin development and a striking degree of synteny and linearity in the EDC locus across a wide range of mammalian (placental and marsupial) genomes suggests an evolutionary conserved regulatory milieu in the EDC. CNEs identified by comparative genomics exhibit dynamic regulatory activity (enhancer or repressor) in differentiating or proliferating conditions. We further demonstrate epidermal-specific, developmental in vivo enhancer activities (DNaseI and transgenic mouse assays) in CNEs, including one within the psoriasis-associated deletion, LCE3C_LCE3B-del. Together, our multidisciplinary study features a network of regulatory elements coordinating developmental EDC gene expression as an unexplored resource for genetic variants in skin diseases.
Figures
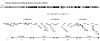
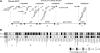


Similar articles
-
On the role of the epidermal differentiation complex in ichthyosis vulgaris, atopic dermatitis and psoriasis.Br J Dermatol. 2007 Sep;157(3):441-9. doi: 10.1111/j.1365-2133.2007.07999.x. Epub 2007 Jun 15. Br J Dermatol. 2007. PMID: 17573887 Review.
-
Deletion of Late Cornified Envelope 3B and 3C genes is not associated with atopic dermatitis.J Invest Dermatol. 2010 Aug;130(8):2057-61. doi: 10.1038/jid.2010.88. Epub 2010 Apr 8. J Invest Dermatol. 2010. PMID: 20376060
-
Lack of association between filaggrin gene mutations and onset of psoriasis in childhood.J Eur Acad Dermatol Venereol. 2013 Jan;27(1):e124-7. doi: 10.1111/j.1468-3083.2011.04403.x. Epub 2011 Dec 20. J Eur Acad Dermatol Venereol. 2013. PMID: 22182180
-
Regulation of the dynamic chromatin architecture of the epidermal differentiation complex is mediated by a c-Jun/AP-1-modulated enhancer.J Invest Dermatol. 2014 Sep;134(9):2371-2380. doi: 10.1038/jid.2014.44. Epub 2014 Jan 27. J Invest Dermatol. 2014. PMID: 24468747 Free PMC article.
-
Potential Role of the Epidermal Differentiation Complex in the Pathogenesis of Psoriasis.Front Biosci (Landmark Ed). 2022 Dec 20;27(12):325. doi: 10.31083/j.fbl2712325. Front Biosci (Landmark Ed). 2022. PMID: 36624944 Review.
Cited by
-
Bayesian model and selection signature analyses reveal risk factors for canine atopic dermatitis.Commun Biol. 2022 Dec 8;5(1):1348. doi: 10.1038/s42003-022-04279-8. Commun Biol. 2022. PMID: 36482174 Free PMC article.
-
Allelic Variants of HLA-C Upstream Region, PSORS1C3, MICA, TNFA and Genes Involved in Epidermal Homeostasis and Barrier Function Influence the Clinical Response to Anti-IL-12/IL-23 Treatment of Patients with Psoriasis.Vaccines (Basel). 2022 Nov 21;10(11):1977. doi: 10.3390/vaccines10111977. Vaccines (Basel). 2022. PMID: 36423071 Free PMC article.
-
High-Quality Genomes of Pangolins: Insights into the Molecular Basis of Scale Formation and Adaption to Myrmecophagous Diet.Mol Biol Evol. 2023 Jan 4;40(1):msac262. doi: 10.1093/molbev/msac262. Mol Biol Evol. 2023. PMID: 36585823 Free PMC article.
-
Liver X receptor-retinoid X receptor (LXR-RXR) heterodimer cistrome reveals coordination of LXR and AP1 signaling in keratinocytes.J Biol Chem. 2011 Apr 22;286(16):14554-63. doi: 10.1074/jbc.M110.165704. Epub 2011 Feb 24. J Biol Chem. 2011. PMID: 21349840 Free PMC article.
-
Tmem79/Matt is the matted mouse gene and is a predisposing gene for atopic dermatitis in human subjects.J Allergy Clin Immunol. 2013 Nov;132(5):1121-9. doi: 10.1016/j.jaci.2013.08.046. Epub 2013 Sep 29. J Allergy Clin Immunol. 2013. PMID: 24084074 Free PMC article.
References
-
- Volz A., Korge B.P., Compton J.G., Ziegler A., Steinert P.M., Mischke D. Physical mapping of a functional cluster of epidermal differentiation genes on chromosome 1q21. Genomics. 1993;18:92–99. - PubMed
-
- Zhao X.P., Elder J.T. Positional cloning of novel skin-specific genes from the human epidermal differentiation complex. Genomics. 1997;45:250–258. - PubMed
-
- Mischke D., Korge B.P., Marenholz I., Volz A., Ziegler A. Genes encoding structural proteins of epidermal cornification and S100 calcium-binding proteins form a gene complex (‘epidermal differentiation complex’) on human chromosome 1q21. J. Invest. Dermatol. 1996;106:989–992. - PubMed
-
- Eckert R.L., Broome A.M., Ruse M., Robinson N., Ryan D., Lee K. S100 proteins in the epidermis. J. Invest. Dermatol. 2004;123:23–33. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

