Combinational soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical autophagosomes with lysosomes
- PMID: 20089838
- PMCID: PMC2836953
- DOI: 10.1091/mbc.e09-08-0693
Combinational soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical autophagosomes with lysosomes
Abstract
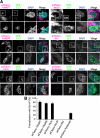
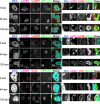
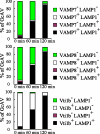
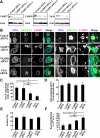
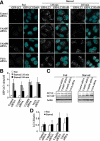
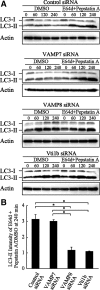
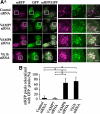
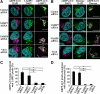
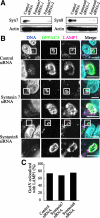
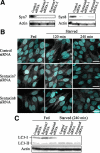
Autophagy plays a crucial role in host defense, termed antimicrobial autophagy (xenophagy), as it functions to degrade intracellular foreign microbial invaders such as group A Streptococcus (GAS). Xenophagosomes undergo a stepwise maturation process consisting of a fusion event with lysosomes, after which the cargoes are degraded. However, the molecular mechanism underlying xenophagosome/lysosome fusion remains unclear. We examined the involvement of endocytic soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) in xenophagosome/lysosome fusion. Confocal microscopic analysis showed that SNAREs, including vesicle-associated membrane protein (VAMP)7, VAMP8, and vesicle transport through interaction with t-SNAREs homologue 1B (Vti1b), colocalized with green fluorescent protein-LC3 in xenophagosomes. Knockdown of Vti1b and VAMP8 with small interfering RNAs disturbed the colocalization of LC3 with lysosomal membrane protein (LAMP)1. The invasive efficiency of GAS into cells was not altered by knockdown of VAMP8 or Vti1b, whereas cellular bactericidal efficiency was significantly diminished, indicating that antimicrobial autophagy was functionally impaired. Knockdown of Vti1b and VAMP8 also disturbed colocalization of LC3 with LAMP1 in canonical autophagy, in which LC3-II proteins were negligibly degraded. In contrast, knockdown of Syntaxin 7 and Syntaxin 8 showed little effect on the autophagic fusion event. These findings strongly suggest that the combinational SNARE proteins VAMP8 and Vti1b mediate the fusion of antimicrobial and canonical autophagosomes with lysosomes, an essential event for autophagic degradation.
Figures
References
-
- Advani R. J., Bae H. R., Bock J. B., Chao D. S., Doung Y. C., Prekeris R., Yoo J. S., Scheller R. H. Seven novel mammalian SNARE proteins localize to distinct membrane compartments. J. Biol. Chem. 1998;273:10317–10324. - PubMed
-
- Bright N. A., Gratian M. J., Luzio J. P. Endocytic delivery to lysosomes mediated by concurrent fusion and kissing events in living cells. Curr. Biol. 2005;15:360–365. - PubMed
-
- Cossart P., Sansonetti P. J. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science. 2004;304:242–248. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

