cAMP-response element-binding protein (CREB) controls MSK1-mediated phosphorylation of histone H3 at the c-fos promoter in vitro
- PMID: 20089855
- PMCID: PMC2843188
- DOI: 10.1074/jbc.M109.057745
cAMP-response element-binding protein (CREB) controls MSK1-mediated phosphorylation of histone H3 at the c-fos promoter in vitro
Abstract
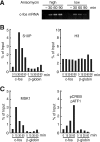
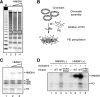
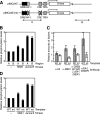
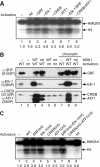
The rapid induction of the c-fos gene correlates with phosphorylations of histone H3 and HMGN1 by mitogen- and stress-activated protein kinases. We have used a cell-free system to dissect the mechanism by which MSK1 phosphorylates histone H3 within the c-fos chromatin. Here, we show that the reconstituted c-fos chromatin presents a strong barrier to histone H3 phosphorylation by MSK1; however, the activators (serum response factor, Elk-1, cAMP-response element-binding protein (CREB), and ATF1) bound on their cognate sites recruit MSK1 to phosphorylate histone H3 at Ser-10 within the chromatin. This activator-dependent phosphorylation of histone H3 is enhanced by HMGN1 and occurs preferentially near the promoter region. Among the four activators, CREB plays a predominant role in MSK1-mediated phosphorylation of histone H3, and the phosphorylation of Ser-133 in CREB is essential for this process. Mutational analyses of MSK1 show that its N-terminal inhibition domain is critical for the kinase to phosphorylate chromatin-embedded histone H3 in a CREB-dependent manner, indicating the presence of an intricate regulatory network for MSK1-mediated phosphorylation of histone H3.
Figures


Similar articles
-
Shear stress-mediated chromatin remodeling provides molecular basis for flow-dependent regulation of gene expression.Circ Res. 2003 Jul 25;93(2):155-61. doi: 10.1161/01.RES.0000080933.82105.29. Epub 2003 Jun 12. Circ Res. 2003. PMID: 12805238
-
Ultraviolet B-induced phosphorylation of histone H3 at serine 28 is mediated by MSK1.J Biol Chem. 2001 Aug 31;276(35):33213-9. doi: 10.1074/jbc.M103973200. Epub 2001 Jul 5. J Biol Chem. 2001. PMID: 11441012
-
Phosphorylation of histone H2A inhibits transcription on chromatin templates.J Biol Chem. 2004 May 21;279(21):21866-72. doi: 10.1074/jbc.M400099200. Epub 2004 Mar 9. J Biol Chem. 2004. PMID: 15010469
-
MSK1 and MSK2 mediate mitogen- and stress-induced phosphorylation of histone H3: a controversy resolved.Sci STKE. 2003 Aug 12;2003(195):PE33. doi: 10.1126/stke.2003.195.pe33. Sci STKE. 2003. PMID: 12915720 Review.
-
Role of the ERK/MSK1 signalling pathway in chromatin remodelling and brain responses to drugs of abuse.J Neurochem. 2009 Mar;108(6):1323-35. doi: 10.1111/j.1471-4159.2009.05879.x. Epub 2009 Jan 12. J Neurochem. 2009. PMID: 19183268 Review.
Cited by
-
Cross-talk between the p38alpha and JNK MAPK pathways mediated by MAP kinase phosphatase-1 determines cellular sensitivity to UV radiation.J Biol Chem. 2010 Aug 20;285(34):25928-40. doi: 10.1074/jbc.M110.117911. Epub 2010 Jun 11. J Biol Chem. 2010. PMID: 20547488 Free PMC article.
-
Cytomegalovirus latency and reactivation: recent insights into an age old problem.Rev Med Virol. 2016 Mar;26(2):75-89. doi: 10.1002/rmv.1862. Epub 2015 Nov 17. Rev Med Virol. 2016. PMID: 26572645 Free PMC article. Review.
-
β-agonists selectively modulate proinflammatory gene expression in skeletal muscle cells via non-canonical nuclear crosstalk mechanisms.PLoS One. 2014 Mar 6;9(6):e90649. doi: 10.1371/journal.pone.0090649. eCollection 2014. PLoS One. 2014. PMID: 24603712 Free PMC article.
-
MSK1 functions as a transcriptional coactivator of p53 in the regulation of p21 gene expression.Exp Mol Med. 2018 Oct 10;50(10):1-12. doi: 10.1038/s12276-018-0160-8. Exp Mol Med. 2018. PMID: 30305627 Free PMC article.
-
Abnormal expression of TFIIIB subunits and RNA Pol III genes is associated with hepatocellular carcinoma.Liver Res. 2017 Sep;1(2):112-120. doi: 10.1016/j.livres.2017.08.005. Epub 2017 Sep 9. Liver Res. 2017. PMID: 29276645 Free PMC article.
References
-
- Price M. A., Hill C., Treisman R. (1996) Philos. Trans. R Soc. Lond. B Biol. Sci. 351, 551–559 - PubMed
-
- Thomson S., Mahadevan L. C., Clayton A. L. (1999) Semin. Cell Dev. Biol. 10, 205–214 - PubMed
-
- Chadee D. N., Hendzel M. J., Tylipski C. P., Allis C. D., Bazett-Jones D. P., Wright J. A., Davie J. R. (1999) J. Biol. Chem. 274, 24914–24920 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

