Structure of the antibiotic resistance factor spectinomycin phosphotransferase from Legionella pneumophila
- PMID: 20089863
- PMCID: PMC2843205
- DOI: 10.1074/jbc.M109.038364
Structure of the antibiotic resistance factor spectinomycin phosphotransferase from Legionella pneumophila
Abstract
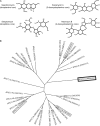
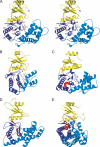
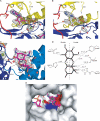
Aminoglycoside phosphotransferases (APHs) constitute a diverse group of enzymes that are often the underlying cause of aminoglycoside resistance in the clinical setting. Several APHs have been extensively characterized, including the elucidation of the three-dimensional structure of two APH(3') isozymes and an APH(2'') enzyme. Although many APHs are plasmid-encoded and are capable of inactivating numerous 2-deoxystreptmaine aminoglycosides with multiple regiospecificity, APH(9)-Ia, isolated from Legionella pneumophila, is an unusual enzyme among the APH family for its chromosomal origin and its specificity for a single non-2-deoxystreptamine aminoglycoside substrate, spectinomycin. We describe here the crystal structures of APH(9)-Ia in its apo form, its binary complex with the nucleotide, AMP, and its ternary complex bound with ADP and spectinomycin. The structures reveal that APH(9)-Ia adopts the bilobal protein kinase-fold, analogous to the APH(3') and APH(2'') enzymes. However, APH(9)-Ia differs significantly from the other two types of APH enzymes in its substrate binding area and that it undergoes a conformation change upon ligand binding. Moreover, kinetic assay experiments indicate that APH(9)-Ia has stringent substrate specificity as it is unable to phosphorylate substrates of choline kinase or methylthioribose kinase despite high structural resemblance. The crystal structures of APH(9)-Ia demonstrate and expand our understanding of the diversity of the APH family, which in turn will facilitate the development of new antibiotics and inhibitors.
Figures
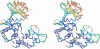
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

