The Escherichia coli PriA helicase specifically recognizes gapped DNA substrates: effect of the two nucleotide-binding sites of the enzyme on the recognition process
- PMID: 20089865
- PMCID: PMC2843218
- DOI: 10.1074/jbc.M109.094789
The Escherichia coli PriA helicase specifically recognizes gapped DNA substrates: effect of the two nucleotide-binding sites of the enzyme on the recognition process
Abstract
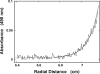
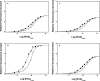
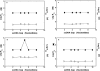
Energetics and specificity of interactions between the Escherichia coli PriA helicase and the gapped DNAs have been studied, using the quantitative fluorescence titration and analytical ultracentrifugation methods. The gap complex has a surprisingly low minimum total site size, corresponding to approximately 7 nucleotides of the single-stranded DNA (ssDNA), as compared with the site size of approximately 20 nucleotides of the enzyme-ssDNA complex. The dramatic difference in stoichiometries indicates that the enzyme predominantly engages the strong DNA-binding subsite in interactions with the gap and assumes a very different orientation in the gap complex, as compared with the complex with the ssDNA. The helicase binds the ssDNA gaps with 4-5 nucleotides with the highest affinity, which is approximately 3 and approximately 2 orders of magnitude larger than the affinities for the ssDNA and double-stranded DNA, respectively. In the gap complex, the protein does not engage in cooperative interactions with the enzyme predominantly associated with the surrounding dsDNA. Binding of nucleoside triphosphate to the strong and weak nucleotide-binding sites of the helicase eliminates the selectivity of the enzyme for the size of the gap, whereas saturation of both sites with ADP leads to amplified affinity for the ssDNA gap containing 5 nucleotides and engagement of an additional protein area in interactions with the nucleic acid.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

