ADAM22, a Kv1 channel-interacting protein, recruits membrane-associated guanylate kinases to juxtaparanodes of myelinated axons
- PMID: 20089912
- PMCID: PMC2811262
- DOI: 10.1523/JNEUROSCI.4661-09.2010
ADAM22, a Kv1 channel-interacting protein, recruits membrane-associated guanylate kinases to juxtaparanodes of myelinated axons
Abstract
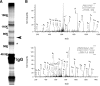
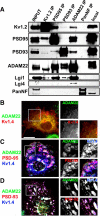
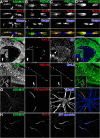
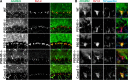
Clustered Kv1 K(+) channels regulate neuronal excitability at juxtaparanodes of myelinated axons, axon initial segments, and cerebellar basket cell terminals (BCTs). These channels are part of a larger protein complex that includes cell adhesion molecules and scaffolding proteins. To identify proteins that regulate assembly, clustering, and/or maintenance of axonal Kv1 channel protein complexes, we immunoprecipitated Kv1.2 alpha subunits, and then used mass spectrometry to identify interacting proteins. We found that a disintegrin and metalloproteinase 22 (ADAM22) is a component of the Kv1 channel complex and that ADAM22 coimmunoprecipitates Kv1.2 and the membrane-associated guanylate kinases (MAGUKs) PSD-93 and PSD-95. When coexpressed with MAGUKs in heterologous cells, ADAM22 and Kv1 channels are recruited into membrane surface clusters. However, coexpression of Kv1.2 with ADAM22 and MAGUKs does not alter channel properties. Among all the known Kv1 channel-interacting proteins, only ADAM22 is found at every site where Kv1 channels are clustered. Analysis of Caspr-null mice showed that, like other previously described juxtaparanodal proteins, disruption of the paranodal junction resulted in redistribution of ADAM22 into paranodal zones. Analysis of Caspr2-, PSD-93-, PSD-95-, and double PSD-93/PSD-95-null mice showed ADAM22 clustering at BCTs requires PSD-95, but ADAM22 clustering at juxtaparanodes requires neither PSD-93 nor PSD-95. In direct contrast, analysis of ADAM22-null mice demonstrated juxtaparanodal clustering of PSD-93 and PSD-95 requires ADAM22, whereas Kv1.2 and Caspr2 clustering is normal in ADAM22-null mice. Thus, ADAM22 is an axonal component of the Kv1 K(+) channel complex that recruits MAGUKs to juxtaparanodes.
Figures



References
-
- Baba H, Akita H, Ishibashi T, Inoue Y, Nakahira K, Ikenaka K. Completion of myelin compaction, but not the attachment of oligodendroglial processes triggers K+ channel clustering. J Neurosci Res. 1999;58:752–764. - PubMed
-
- Berghs S, Aggujaro D, Dirkx R, Jr, Maksimova E, Stabach P, Hermel JM, Zhang JP, Philbrick W, Slepnev V, Ort T, Solimena M. betaIV spectrin, a new spectrin localized at axon initial segments and nodes of Ranvier in the central and peripheral nervous system. J Cell Biol. 2000;151:985–1002. - PMC - PubMed
-
- Bermingham JR, Jr, Shearin H, Pennington J, O'Moore J, Jaegle M, Driegen S, van Zon A, Darbas A, Ozkaynak E, Ryu EJ, Milbrandt J, Meijer D. The claw paw mutation reveals a role for Lgi4 in peripheral nerve development. Nat Neurosci. 2006;9:76–84. - PubMed
-
- Chabrol E, Gourfinkel-An I, Scheffer IE, Picard F, Couarch P, Berkovic SF, McMahon JM, Bajaj N, Mota-Vieira L, Mota R, Trouillard O, Depienne C, Baulac M, LeGuern E, Baulac S, et al. Absence of mutations in the LGI1 receptor ADAM22 gene in autosomal dominant lateral temporal epilepsy. Epilepsy Res. 2007;76:41–48. - PubMed
-
- Clauser KR, Baker P, Burlingame AL. Role of accurate mass measurement (±10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal Chem. 1999;71:2871–2882. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U24 NS050606/NS/NINDS NIH HHS/United States
- NS034383/NS/NINDS NIH HHS/United States
- RR019934/RR/NCRR NIH HHS/United States
- S10 RR019934/RR/NCRR NIH HHS/United States
- U24NS050606/NS/NINDS NIH HHS/United States
- NS044916/NS/NINDS NIH HHS/United States
- P41 RR001614/RR/NCRR NIH HHS/United States
- R37 NS034383/NS/NINDS NIH HHS/United States
- R37 NS044916/NS/NINDS NIH HHS/United States
- R01 NS044916/NS/NINDS NIH HHS/United States
- R01 NS034383/NS/NINDS NIH HHS/United States
- P41RR001614/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
