Mutational robustness can facilitate adaptation
- PMID: 20090752
- PMCID: PMC3071712
- DOI: 10.1038/nature08694
Mutational robustness can facilitate adaptation
Abstract
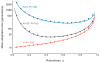
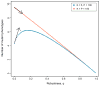
Robustness seems to be the opposite of evolvability. If phenotypes are robust against mutation, we might expect that a population will have difficulty adapting to an environmental change, as several studies have suggested. However, other studies contend that robust organisms are more adaptable. A quantitative understanding of the relationship between robustness and evolvability will help resolve these conflicting reports and will clarify outstanding problems in molecular and experimental evolution, evolutionary developmental biology and protein engineering. Here we demonstrate, using a general population genetics model, that mutational robustness can either impede or facilitate adaptation, depending on the population size, the mutation rate and the structure of the fitness landscape. In particular, neutral diversity in a robust population can accelerate adaptation as long as the number of phenotypes accessible to an individual by mutation is smaller than the total number of phenotypes in the fitness landscape. These results provide a quantitative resolution to a significant ambiguity in evolutionary theory.
Conflict of interest statement
Figures
Similar articles
-
Recombination accelerates adaptation on a large-scale empirical fitness landscape in HIV-1.PLoS Genet. 2014 Jun 26;10(6):e1004439. doi: 10.1371/journal.pgen.1004439. eCollection 2014 Jun. PLoS Genet. 2014. PMID: 24967626 Free PMC article.
-
Evolutionary rescue from extinction is contingent on a lower rate of environmental change.Nature. 2013 Feb 28;494(7438):463-7. doi: 10.1038/nature11879. Epub 2013 Feb 10. Nature. 2013. PMID: 23395960
-
Robustness and evolvability: a paradox resolved.Proc Biol Sci. 2008 Jan 7;275(1630):91-100. doi: 10.1098/rspb.2007.1137. Proc Biol Sci. 2008. PMID: 17971325 Free PMC article.
-
How evolutionary systems biology will help understand adaptive landscapes and distributions of mutational effects.Adv Exp Med Biol. 2012;751:399-410. doi: 10.1007/978-1-4614-3567-9_18. Adv Exp Med Biol. 2012. PMID: 22821468 Review.
-
The role of robustness in phenotypic adaptation and innovation.Proc Biol Sci. 2012 Apr 7;279(1732):1249-58. doi: 10.1098/rspb.2011.2293. Epub 2012 Jan 4. Proc Biol Sci. 2012. PMID: 22217723 Free PMC article. Review.
Cited by
-
Robustness, evolvability, and the logic of genetic regulation.Artif Life. 2014 Winter;20(1):111-26. doi: 10.1162/ARTL_a_00099. Epub 2013 Feb 1. Artif Life. 2014. PMID: 23373974 Free PMC article.
-
Chaperones, Canalization, and Evolution of Animal Forms.Int J Mol Sci. 2018 Oct 4;19(10):3029. doi: 10.3390/ijms19103029. Int J Mol Sci. 2018. PMID: 30287767 Free PMC article. Review.
-
A Family of Fitness Landscapes Modeled through Gene Regulatory Networks.Entropy (Basel). 2022 Apr 29;24(5):622. doi: 10.3390/e24050622. Entropy (Basel). 2022. PMID: 35626507 Free PMC article.
-
A systematic survey of an intragenic epistatic landscape.Mol Biol Evol. 2015 Jan;32(1):229-38. doi: 10.1093/molbev/msu301. Epub 2014 Nov 3. Mol Biol Evol. 2015. PMID: 25371431 Free PMC article.
-
Mutational robustness and resilience of a replicative cis-element of RNA virus: Promiscuity, limitations, relevance.RNA Biol. 2015;12(12):1338-54. doi: 10.1080/15476286.2015.1100794. RNA Biol. 2015. PMID: 26488412 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

