Associating genes and protein complexes with disease via network propagation
- PMID: 20090828
- PMCID: PMC2797085
- DOI: 10.1371/journal.pcbi.1000641
Associating genes and protein complexes with disease via network propagation
Abstract
A fundamental challenge in human health is the identification of disease-causing genes. Recently, several studies have tackled this challenge via a network-based approach, motivated by the observation that genes causing the same or similar diseases tend to lie close to one another in a network of protein-protein or functional interactions. However, most of these approaches use only local network information in the inference process and are restricted to inferring single gene associations. Here, we provide a global, network-based method for prioritizing disease genes and inferring protein complex associations, which we call PRINCE. The method is based on formulating constraints on the prioritization function that relate to its smoothness over the network and usage of prior information. We exploit this function to predict not only genes but also protein complex associations with a disease of interest. We test our method on gene-disease association data, evaluating both the prioritization achieved and the protein complexes inferred. We show that our method outperforms extant approaches in both tasks. Using data on 1,369 diseases from the OMIM knowledgebase, our method is able (in a cross validation setting) to rank the true causal gene first for 34% of the diseases, and infer 139 disease-related complexes that are highly coherent in terms of the function, expression and conservation of their member proteins. Importantly, we apply our method to study three multi-factorial diseases for which some causal genes have been found already: prostate cancer, alzheimer and type 2 diabetes mellitus. PRINCE's predictions for these diseases highly match the known literature, suggesting several novel causal genes and protein complexes for further investigation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
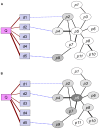
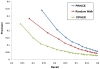
 proteins for various values of
proteins for various values of  .
.
References
-
- Oti M, Brunner HG. The modular nature of genetic diseases. Clinical Genetics. 2007;71:1–11. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

