beta-Neurexin is a ligand for the Staphylococcus aureus MSCRAMM SdrC
- PMID: 20090838
- PMCID: PMC2800189
- DOI: 10.1371/journal.ppat.1000726
beta-Neurexin is a ligand for the Staphylococcus aureus MSCRAMM SdrC
Abstract
Gram-positive bacteria contain a family of surface proteins that are covalently anchored to the cell wall of the organism. These cell-wall anchored (CWA) proteins appear to play key roles in the interactions between pathogenic organisms and the host. A subfamily of the CWA has a common structural organization with multiple domains adopting characteristic IgG-like folds. The identified microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) belong to this subfamily, as does SdrC from S. aureus. However, an interactive host ligand for the putative MSCRAMM SdrC was not previously identified. We have screened a phage display peptide library and identified a peptide sequence found in beta-neurexin that binds SdrC. A synthetic peptide corresponding to the identified sequence as well as a recombinant form of the beta-neurexin 1 exodomain binds SdrC with high affinity and specificity. Furthermore, expression of SdrC on bacteria greatly enhances microbial adherence to cultured mammalian cells expressing beta-neurexin on their surface. Taken together, our experimental results demonstrate that beta-neurexin is a ligand for SdrC. This interaction involves a specific sequence located in the N-terminal region of the mammalian protein and the N(2)N(3) domain of the MSCRAMM. The fact that these two proteins interact when expressed on the appropriate cells demonstrates the functionality of the interaction. Possible implications of this interaction are discussed.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


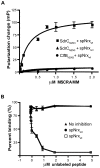

Similar articles
-
Clumping factor B, a fibrinogen-binding MSCRAMM (microbial surface components recognizing adhesive matrix molecules) adhesin of Staphylococcus aureus, also binds to the tail region of type I cytokeratin 10.J Biol Chem. 2004 Dec 3;279(49):50691-9. doi: 10.1074/jbc.M408713200. Epub 2004 Sep 22. J Biol Chem. 2004. PMID: 15385531
-
Multiple specificities of the staphylococcal and streptococcal fibronectin-binding microbial surface components recognizing adhesive matrix molecules.Eur J Biochem. 1998 Dec 1;258(2):897-905. doi: 10.1046/j.1432-1327.1998.2580897.x. Eur J Biochem. 1998. PMID: 9874261
-
Fibrinogen is a ligand for the Staphylococcus aureus microbial surface components recognizing adhesive matrix molecules (MSCRAMM) bone sialoprotein-binding protein (Bbp).J Biol Chem. 2011 Aug 26;286(34):29797-805. doi: 10.1074/jbc.M110.214981. Epub 2011 Jun 3. J Biol Chem. 2011. PMID: 21642438 Free PMC article.
-
Surface Proteins of Staphylococcus aureus.Microbiol Spectr. 2019 Jul;7(4):10.1128/microbiolspec.gpp3-0046-2018. doi: 10.1128/microbiolspec.GPP3-0046-2018. Microbiol Spectr. 2019. PMID: 31267926 Free PMC article. Review.
-
The MSCRAMM Family of Cell-Wall-Anchored Surface Proteins of Gram-Positive Cocci.Trends Microbiol. 2019 Nov;27(11):927-941. doi: 10.1016/j.tim.2019.06.007. Epub 2019 Jul 30. Trends Microbiol. 2019. PMID: 31375310 Review.
Cited by
-
Adhesive Virulence Factors of Staphylococcus aureus Resist Digestion by Coagulation Proteases Thrombin and Plasmin.ACS Bio Med Chem Au. 2022 Dec 21;2(6):586-599. doi: 10.1021/acsbiomedchemau.2c00042. Epub 2022 Sep 2. ACS Bio Med Chem Au. 2022. PMID: 36573096 Free PMC article.
-
Mechanism of antibacterial and antibiofilm of thiazolidinone derivative TD-H2-A against Staphylococcus aureus.Sci Rep. 2025 Mar 26;15(1):10380. doi: 10.1038/s41598-025-94571-4. Sci Rep. 2025. PMID: 40140486 Free PMC article.
-
Structures of SdrD from Staphylococcus aureus reveal the molecular mechanism of how the cell surface receptors recognize their ligands.Protein Cell. 2013 Apr;4(4):277-85. doi: 10.1007/s13238-013-3009-x. Epub 2013 Apr 3. Protein Cell. 2013. PMID: 23549613 Free PMC article.
-
Characterization of transcription within sdr region of Staphylococcus aureus.Antonie Van Leeuwenhoek. 2011 Feb;99(2):409-16. doi: 10.1007/s10482-010-9476-7. Epub 2010 Jun 24. Antonie Van Leeuwenhoek. 2011. PMID: 20571861 Free PMC article.
-
SdrC induces staphylococcal biofilm formation through a homophilic interaction.Mol Microbiol. 2014 Oct;94(1):172-85. doi: 10.1111/mmi.12750. Epub 2014 Sep 3. Mol Microbiol. 2014. PMID: 25115812 Free PMC article.
References
-
- Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. - PubMed
-
- Clarke SR, Foster SJ. Surface adhesins of Staphylococcus aureus. Adv Microb Physiol. 2006;51:187–224. - PubMed
-
- Patti JM, Allen BL, McGavin MJ, Hook M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol. 1994;48:585–617. - PubMed
-
- Patti JM, Hook M. Microbial adhesins recognizing extracellular matrix macromolecules. Curr Opin Cell Biol. 1994;6:752–758. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

