Derivation of Escherichia coli O157:H7 from its O55:H7 precursor
- PMID: 20090843
- PMCID: PMC2806823
- DOI: 10.1371/journal.pone.0008700
Derivation of Escherichia coli O157:H7 from its O55:H7 precursor
Abstract
There are 29 E. coli genome sequences available, mostly related to studies of species diversity or mode of pathogenicity, including two genomes of the well-known O157:H7 clone. However, there have been no genome studies of closely related clones aimed at exposing the details of evolutionary change. Here we sequenced the genome of an O55:H7 strain, closely related to the major pathogenic O157:H7 clone, with published genome sequences, and undertook comparative genomic and proteomic analysis. We were able to allocate most differences between the genomes to individual mutations, recombination events, or lateral gene transfer events, in specific lineages. Major differences include a type II secretion system present only in the O55:H7 chromosome, fewer type III secretion system effectors in O55:H7, and 19 phage genomes or phagelike elements in O55:H7 compared to 23 in O157:H7, with only three common to both. Many other changes were found in both O55:H7 and O157:H7 lineages, but in general there has been more change in the O157:H7 lineages. For example, we found 50% more synonymous mutational substitutions in O157:H7 compared to O55:H7. The two strains also diverged at the proteomic level. Mutational synonymous SNPs were used to estimate a divergence time of 400 years using a new clock rate, in contrast to 14,000 to 70,000 years using the traditional clock rates. The same approaches were applied to three closely related extraintestinal pathogenic E. coli genomes, and similar levels of mutation and recombination were found. This study revealed for the first time the full range of events involved in the evolution of the O157:H7 clone from its O55:H7 ancestor, and suggested that O157:H7 arose quite recently. Our findings also suggest that E. coli has a much lower frequency of recombination relative to mutation than was observed in a comparable study of a Vibrio cholerae lineage.
Conflict of interest statement
Figures

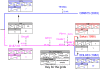
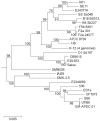

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

