Crystal structure of the S. solfataricus archaeal exosome reveals conformational flexibility in the RNA-binding ring
- PMID: 20090900
- PMCID: PMC2806925
- DOI: 10.1371/journal.pone.0008739
Crystal structure of the S. solfataricus archaeal exosome reveals conformational flexibility in the RNA-binding ring
Abstract
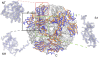
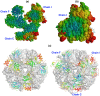
Background: The exosome complex is an essential RNA 3'-end processing and degradation machinery. In archaeal organisms, the exosome consists of a catalytic ring and an RNA-binding ring, both of which were previously reported to assume three-fold symmetry.
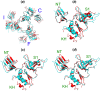
Methodology/principal findings: Here we report an asymmetric 2.9 A Sulfolobus solfataricus archaeal exosome structure in which the three-fold symmetry is broken due to combined rigid body and thermal motions mainly within the RNA-binding ring. Since increased conformational flexibility was also observed in the RNA-binding ring of the related bacterial PNPase, we speculate that this may reflect an evolutionarily conserved mechanism to accommodate diverse RNA substrates for degradation.
Conclusion/significance: This study clearly shows the dynamic structures within the RNA-binding domains, which provides additional insights on mechanism of asymmetric RNA binding and processing.
Conflict of interest statement
Figures

Similar articles
-
Expression, reconstitution, and structure of an archaeal RNA degrading exosome.Methods Enzymol. 2008;447:417-35. doi: 10.1016/S0076-6879(08)02220-9. Methods Enzymol. 2008. PMID: 19161854
-
The archaeal exosome.Adv Exp Med Biol. 2011;702:29-38. doi: 10.1007/978-1-4419-7841-7_3. Adv Exp Med Biol. 2011. PMID: 21713675
-
Nop5 interacts with the archaeal RNA exosome.FEBS Lett. 2017 Dec;591(24):4039-4048. doi: 10.1002/1873-3468.12915. Epub 2017 Nov 28. FEBS Lett. 2017. PMID: 29159940
-
Structure and function of the archaeal exosome.Wiley Interdiscip Rev RNA. 2014 Sep-Oct;5(5):623-35. doi: 10.1002/wrna.1234. Epub 2014 Apr 30. Wiley Interdiscip Rev RNA. 2014. PMID: 24789718 Review.
-
The archaeal exosome.Adv Exp Med Biol. 2010;702:29-38. Adv Exp Med Biol. 2010. PMID: 21618872 Review.
Cited by
-
Enzymatic Analysis of Reconstituted Archaeal Exosomes.Methods Mol Biol. 2020;2062:63-79. doi: 10.1007/978-1-4939-9822-7_4. Methods Mol Biol. 2020. PMID: 31768972
-
Crystal structure of a 9-subunit archaeal exosome in pre-catalytic states of the phosphorolytic reaction.Archaea. 2012;2012:721869. doi: 10.1155/2012/721869. Epub 2012 Dec 20. Archaea. 2012. PMID: 23319881 Free PMC article.
-
RNA degradation by the plant RNA exosome involves both phosphorolytic and hydrolytic activities.Nat Commun. 2017 Dec 18;8(1):2162. doi: 10.1038/s41467-017-02066-2. Nat Commun. 2017. PMID: 29255150 Free PMC article.
-
Structure and Activities of the Eukaryotic RNA Exosome.Enzymes. 2012;31:53-75. doi: 10.1016/B978-0-12-404740-2.00003-3. Epub 2012 Sep 29. Enzymes. 2012. PMID: 27166440 Free PMC article.
-
RNA channelling by the eukaryotic exosome.EMBO Rep. 2010 Dec;11(12):936-42. doi: 10.1038/embor.2010.164. Epub 2010 Nov 12. EMBO Rep. 2010. PMID: 21072061 Free PMC article.
References
-
- Butler JS. The yin and yang of the exosome. Trends Cell Biol. 2002;12:90–96. S0962892401022255 [pii] - PubMed
-
- Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–539. nrm1964 [pii] 10.1038/nrm1964 [doi] - PubMed
-
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nature Structural & Molecular Biology. 2004;11:121–127. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

