Diversity of Haloquadratum and other haloarchaea in three, geographically distant, Australian saltern crystallizer ponds
- PMID: 20091074
- PMCID: PMC2832888
- DOI: 10.1007/s00792-009-0295-6
Diversity of Haloquadratum and other haloarchaea in three, geographically distant, Australian saltern crystallizer ponds
Abstract
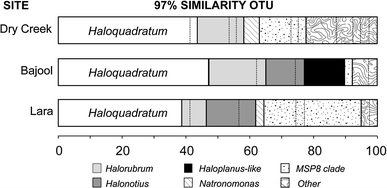
Haloquadratum walsbyi is frequently a dominant member of the microbial communities in hypersaline waters. 16S rRNA gene sequences indicate that divergence within this species is very low but relatively few sites have been examined, particularly in the southern hemisphere. The diversity of Haloquadratum was examined in three coastal, but geographically distant saltern crystallizer ponds in Australia, using both culture-independent and culture-dependent methods. Two 97%-OTU, comprising Haloquadratum- and Halorubrum-related sequences, were shared by all three sites, with the former OTU representing about 40% of the sequences recovered at each site. Sequences 99.5% identical to that of Hqr. walsbyi C23(T) were present at all three sites and, overall, 98% of the Haloquadratum-related sequences displayed <or=2% divergence from that of the type strain. While haloarchaeal diversity at each site was relatively low (9-16 OTUs), seven phylogroups (clones and/or isolates) and 4 different clones showed <or=90% sequence identity to classified taxa, and appear to represent novel genera. Six of these branched together in phylogenetic tree reconstructions, forming a clade (MSP8-clade) whose members were only distantly related to classified taxa. Such sequences have only rarely been previously detected but were found at all three Australian crystallizers.
Figures



References
-
- Antón J, Oren A, Benlloch S, Rodríguez-Valera F, Amann R, Rossello-Mora R. Salinibacter ruber gen. nov., sp. nov., a novel, extremely halophilic member of the Bacteria from saltern crystallizer ponds. Int J Syst Evol Microbiol. 2002;52:485–491. - PubMed
-
- Benlloch S, López-López A, Casamayor EO, Øvreås L, Goddard V, Daae FL, Smerdon G, Massana R, Joint I, Thingstad F, Pedrós-Alió C, Rodríguez-Valera F. Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environ Microbiol. 2002;4:349–360. doi: 10.1046/j.1462-2920.2002.00306.x. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

