Nucleocytoplasmic functions of the PDZ-LIM protein family: new insights into organ development
- PMID: 20091751
- PMCID: PMC3010972
- DOI: 10.1002/bies.200900148
Nucleocytoplasmic functions of the PDZ-LIM protein family: new insights into organ development
Abstract
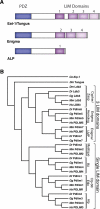
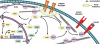
Recent work on the PDZ-LIM protein family has revealed that it has important activities at the cellular level, mediating signals between the nucleus and the cytoskeleton, with significant impact on organ development. We review and integrate current knowledge about the PDZ-LIM protein family and propose a new functional role, sequestering nuclear factors in the cytoplasm. Characterized by their PDZ and LIM domains, the PDZ-LIM family is comprised of evolutionarily conserved proteins found throughout the animal kingdom, from worms to humans. Combining two functional domains in one protein, PDZ-LIM proteins have wide-ranging and multi-compartmental cell functions during development and homeostasis. In contrast, misregulation can lead to cancer formation and progression. New emerging roles include interactions with integrins, T-box transcription factors, and receptor tyrosine kinases. Facilitating the assembly of protein complexes, PDZ-LIM proteins can act as signal modulators, influence actin dynamics, regulate cell architecture, and control gene transcription.
Figures


Similar articles
-
Characterization of the Enigma family in zebrafish.Dev Dyn. 2007 Nov;236(11):3144-54. doi: 10.1002/dvdy.21346. Dev Dyn. 2007. PMID: 17937393
-
ENH, containing PDZ and LIM domains, heart/skeletal muscle-specific protein, associates with cytoskeletal proteins through the PDZ domain.Biochem Biophys Res Commun. 2000 Jun 7;272(2):505-12. doi: 10.1006/bbrc.2000.2787. Biochem Biophys Res Commun. 2000. PMID: 10833443
-
The tumor suppressor Scrib selectively interacts with specific members of the zyxin family of proteins.FEBS Lett. 2005 Sep 12;579(22):5061-8. doi: 10.1016/j.febslet.2005.08.012. FEBS Lett. 2005. PMID: 16137684
-
LIM proteins: association with the actin cytoskeleton.Protoplasma. 2002 Feb;219(1-2):1-12. doi: 10.1007/s007090200000. Protoplasma. 2002. PMID: 11926060 Review.
-
The diverse biofunctions of LIM domain proteins: determined by subcellular localization and protein-protein interaction.Biol Cell. 2007 Sep;99(9):489-502. doi: 10.1042/BC20060126. Biol Cell. 2007. PMID: 17696879 Review.
Cited by
-
The emerin-binding transcription factor Lmo7 is regulated by association with p130Cas at focal adhesions.PeerJ. 2013 Aug 20;1:e134. doi: 10.7717/peerj.134. eCollection 2013. PeerJ. 2013. PMID: 24010014 Free PMC article.
-
Identifying cell types from spatially referenced single-cell expression datasets.PLoS Comput Biol. 2014 Sep 25;10(9):e1003824. doi: 10.1371/journal.pcbi.1003824. eCollection 2014 Sep. PLoS Comput Biol. 2014. PMID: 25254363 Free PMC article.
-
Loss of microRNA-17∼92 in smooth muscle cells attenuates experimental pulmonary hypertension via induction of PDZ and LIM domain 5.Am J Respir Crit Care Med. 2015 Mar 15;191(6):678-92. doi: 10.1164/rccm.201405-0941OC. Am J Respir Crit Care Med. 2015. PMID: 25647182 Free PMC article.
-
Born to run: creating the muscle fiber.Curr Opin Cell Biol. 2010 Oct;22(5):566-74. doi: 10.1016/j.ceb.2010.08.009. Curr Opin Cell Biol. 2010. PMID: 20817426 Free PMC article. Review.
-
Top-down Mass Spectrometry of Sarcomeric Protein Post-translational Modifications from Non-human Primate Skeletal Muscle.J Am Soc Mass Spectrom. 2019 Dec;30(12):2460-2469. doi: 10.1007/s13361-019-02139-0. Epub 2019 Mar 4. J Am Soc Mass Spectrom. 2019. PMID: 30834509 Free PMC article.
References
-
- Wu R, Durick K, Songyang Z, Cantley LC, Taylor SS, Gill GN. Specificity of LIM domain interactions with receptor tyrosine kinases. J Biol Chem. 1996;271(27):15934–41. - PubMed
-
- Durick K, Wu RY, Gill GN, Taylor SS. Mitogenic signaling by Ret/ptc2 requires association with enigma via a LIM domain. J Biol Chem. 1996;271(22):12691–4. - PubMed
-
- Kuroda S, Tokunaga C, Kiyohara Y, Higuchi O, Konishi H, Mizuno K, Gill GN, Kikkawa U. Protein-protein interaction of zinc finger LIM domains with protein kinase C. J Biol Chem. 1996;271(49):31029–32. - PubMed
-
- Zhou Q, Ruiz-Lozano P, Martone ME, Chen J. Cypher, a striated muscle-restricted PDZ and LIM domain-containing protein, binds to alpha-actinin-2 and protein kinase C. J Biol Chem. 1999;274(28):19807–13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

