A comparative approach for the investigation of biological information processing: an examination of the structure and function of computer hard drives and DNA
- PMID: 20092652
- PMCID: PMC2829000
- DOI: 10.1186/1742-4682-7-3
A comparative approach for the investigation of biological information processing: an examination of the structure and function of computer hard drives and DNA
Abstract
Background: The robust storage, updating and utilization of information are necessary for the maintenance and perpetuation of dynamic systems. These systems can exist as constructs of metal-oxide semiconductors and silicon, as in a digital computer, or in the "wetware" of organic compounds, proteins and nucleic acids that make up biological organisms. We propose that there are essential functional properties of centralized information-processing systems; for digital computers these properties reside in the computer's hard drive, and for eukaryotic cells they are manifest in the DNA and associated structures.
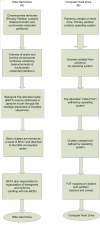
Methods: Presented herein is a descriptive framework that compares DNA and its associated proteins and sub-nuclear structure with the structure and function of the computer hard drive. We identify four essential properties of information for a centralized storage and processing system: (1) orthogonal uniqueness, (2) low level formatting, (3) high level formatting and (4) translation of stored to usable form. The corresponding aspects of the DNA complex and a computer hard drive are categorized using this classification. This is intended to demonstrate a functional equivalence between the components of the two systems, and thus the systems themselves.
Results: Both the DNA complex and the computer hard drive contain components that fulfill the essential properties of a centralized information storage and processing system. The functional equivalence of these components provides insight into both the design process of engineered systems and the evolved solutions addressing similar system requirements. However, there are points where the comparison breaks down, particularly when there are externally imposed information-organizing structures on the computer hard drive. A specific example of this is the imposition of the File Allocation Table (FAT) during high level formatting of the computer hard drive and the subsequent loading of an operating system (OS). Biological systems do not have an external source for a map of their stored information or for an operational instruction set; rather, they must contain an organizational template conserved within their intra-nuclear architecture that "manipulates" the laws of chemistry and physics into a highly robust instruction set. We propose that the epigenetic structure of the intra-nuclear environment and the non-coding RNA may play the roles of a Biological File Allocation Table (BFAT) and biological operating system (Bio-OS) in eukaryotic cells.
Conclusions: The comparison of functional and structural characteristics of the DNA complex and the computer hard drive leads to a new descriptive paradigm that identifies the DNA as a dynamic storage system of biological information. This system is embodied in an autonomous operating system that inductively follows organizational structures, data hierarchy and executable operations that are well understood in the computer science industry. Characterizing the "DNA hard drive" in this fashion can lead to insights arising from discrepancies in the descriptive framework, particularly with respect to positing the role of epigenetic processes in an information-processing context. Further expansions arising from this comparison include the view of cells as parallel computing machines and a new approach towards characterizing cellular control systems.
Figures





Similar articles
-
Planning Implications Related to Sterilization-Sensitive Science Investigations Associated with Mars Sample Return (MSR).Astrobiology. 2022 Jun;22(S1):S112-S164. doi: 10.1089/AST.2021.0113. Epub 2022 May 19. Astrobiology. 2022. PMID: 34904892
-
A DNA network as an information processing system.Int J Mol Sci. 2012;13(4):5125-5137. doi: 10.3390/ijms13045125. Epub 2012 Apr 23. Int J Mol Sci. 2012. PMID: 22606034 Free PMC article.
-
Towards human-computer synergetic analysis of large-scale biological data.BMC Bioinformatics. 2013;14 Suppl 14(Suppl 14):S10. doi: 10.1186/1471-2105-14-S14-S10. Epub 2013 Oct 9. BMC Bioinformatics. 2013. PMID: 24267485 Free PMC article.
-
Preliminary Planning for Mars Sample Return (MSR) Curation Activities in a Sample Receiving Facility (SRF).Astrobiology. 2022 Jun;22(S1):S57-S80. doi: 10.1089/AST.2021.0105. Epub 2022 May 19. Astrobiology. 2022. PMID: 34904890 Review.
-
On eukaryotic intelligence: signaling system's guidance in the evolution of multicellular organization.Biosystems. 2013 Oct;114(1):8-24. doi: 10.1016/j.biosystems.2013.06.005. Epub 2013 Jul 12. Biosystems. 2013. PMID: 23850535 Review.
Cited by
-
Dichotomy in the definition of prescriptive information suggests both prescribed data and prescribed algorithms: biosemiotics applications in genomic systems.Theor Biol Med Model. 2012 Mar 14;9:8. doi: 10.1186/1742-4682-9-8. Theor Biol Med Model. 2012. PMID: 22413926 Free PMC article. Review.
-
A Systems Engineering Perspective on Homeostasis and Disease.Front Bioeng Biotechnol. 2013 Sep 9;1:6. doi: 10.3389/fbioe.2013.00006. eCollection 2013. Front Bioeng Biotechnol. 2013. PMID: 25022216 Free PMC article. Review.
-
Is life unique?Life (Basel). 2011 Dec 30;2(1):106-34. doi: 10.3390/life2010106. Life (Basel). 2011. PMID: 25382119 Free PMC article. Review.
-
Characteristics of Gracilariopsis lemaneiformis hydrocolloids and their effects on intestine PPAR signaling and liver lipid metabolism in Oreochromis niloticus: A multiomics analysis.Heliyon. 2024 Nov 20;10(23):e40416. doi: 10.1016/j.heliyon.2024.e40416. eCollection 2024 Dec 15. Heliyon. 2024. PMID: 39669144 Free PMC article.
-
The Role of Cell Membrane Information Reception, Processing, and Communication in the Structure and Function of Multicellular Tissue.Int J Mol Sci. 2019 Jul 24;20(15):3609. doi: 10.3390/ijms20153609. Int J Mol Sci. 2019. PMID: 31344783 Free PMC article. Review.
References
-
- Regev A, Shapiro EY, (Eds) Cells as Computation. Proceedings of the First International, Workshop on Computational Methods in Systems Biology. London, UK: Springer-Verlag; 2003.
-
- Mueller S. Upgrading and Repairing PC's. 18. USA: Que Publishing; 2007. Magnetic Storage Principles; pp. 637–706.
-
- KozierokThe PC Guide. 1997. pp. 1–7.http://www.pcguide.com
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

