The Swedish new variant of Chlamydia trachomatis: genome sequence, morphology, cell tropism and phenotypic characterization
- PMID: 20093289
- PMCID: PMC3541825
- DOI: 10.1099/mic.0.036830-0
The Swedish new variant of Chlamydia trachomatis: genome sequence, morphology, cell tropism and phenotypic characterization
Abstract
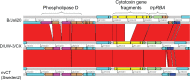
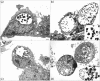
Chlamydia trachomatis is a major cause of bacterial sexually transmitted infections worldwide. In 2006, a new variant of C. trachomatis (nvCT), carrying a 377 bp deletion within the plasmid, was reported in Sweden. This deletion included the targets used by the commercial diagnostic systems from Roche and Abbott. The nvCT is clonal (serovar/genovar E) and it spread rapidly in Sweden, undiagnosed by these systems. The degree of spread may also indicate an increased biological fitness of nvCT. The aims of this study were to describe the genome of nvCT, to compare the nvCT genome to all available C. trachomatis genome sequences and to investigate the biological properties of nvCT. An early nvCT isolate (Sweden2) was analysed by genome sequencing, growth kinetics, microscopy, cell tropism assay and antimicrobial susceptibility testing. It was compared with relevant C. trachomatis isolates, including a similar serovar E C. trachomatis wild-type strain that circulated in Sweden prior to the initially undetected expansion of nvCT. The nvCT genome does not contain any major genetic polymorphisms - the genes for central metabolism, development cycle and virulence are conserved - or phenotypic characteristics that indicate any altered biological fitness. This is supported by the observations that the nvCT and wild-type C. trachomatis infections are very similar in terms of epidemiological distribution, and that differences in clinical signs are only described, in one study, in women. In conclusion, the nvCT does not appear to have any altered biological fitness. Therefore, the rapid transmission of nvCT in Sweden was due to the strong diagnostic selective advantage and its introduction into a high-frequency transmitting population.
Figures

Similar articles
-
The Swedish new variant of Chlamydia trachomatis.Curr Opin Infect Dis. 2011 Feb;24(1):62-9. doi: 10.1097/QCO.0b013e32834204d5. Curr Opin Infect Dis. 2011. PMID: 21157332 Review.
-
Rise and fall of the new variant of Chlamydia trachomatis in Sweden: mathematical modelling study.Sex Transm Infect. 2020 Aug;96(5):375-379. doi: 10.1136/sextrans-2019-054057. Epub 2019 Oct 5. Sex Transm Infect. 2020. PMID: 31586947 Free PMC article.
-
Ten years transmission of the new variant of Chlamydia trachomatis in Sweden: prevalence of infections and associated complications.Sex Transm Infect. 2018 Mar;94(2):100-104. doi: 10.1136/sextrans-2016-052992. Epub 2017 Jul 19. Sex Transm Infect. 2018. PMID: 28724744 Free PMC article.
-
Prevalence trends in Sweden for the new variant of Chlamydia trachomatis.Clin Microbiol Infect. 2011 May;17(5):683-9. doi: 10.1111/j.1469-0691.2010.03305.x. Clin Microbiol Infect. 2011. PMID: 20636428
-
Genomic features beyond Chlamydia trachomatis phenotypes: what do we think we know?Infect Genet Evol. 2013 Jun;16:392-400. doi: 10.1016/j.meegid.2013.03.018. Epub 2013 Mar 21. Infect Genet Evol. 2013. PMID: 23523596 Review.
Cited by
-
Study of gonococcal and chlamydial urethritis: Old culprits with a new story.J Family Med Prim Care. 2022 Sep;11(9):5551-5555. doi: 10.4103/jfmpc.jfmpc_10_21. Epub 2022 Oct 14. J Family Med Prim Care. 2022. PMID: 36505551 Free PMC article.
-
Host-Pathogen Interactions of Chlamydia trachomatis in Porcine Oviduct Epithelial Cells.Pathogens. 2021 Oct 1;10(10):1270. doi: 10.3390/pathogens10101270. Pathogens. 2021. PMID: 34684219 Free PMC article.
-
A Coming of Age Story: Chlamydia in the Post-Genetic Era.Infect Immun. 2015 Dec 14;84(3):612-21. doi: 10.1128/IAI.01186-15. Infect Immun. 2015. PMID: 26667838 Free PMC article. Review.
-
Whole-genome sequences of Chlamydia trachomatis directly from clinical samples without culture.Genome Res. 2013 May;23(5):855-66. doi: 10.1101/gr.150037.112. Epub 2013 Mar 22. Genome Res. 2013. PMID: 23525359 Free PMC article.
-
Letter to the editor: Chlamydia trachomatis samples testing falsely negative in the Aptima Combo 2 test in Finland, 2019.Euro Surveill. 2019 Jun;24(24):1900354. doi: 10.2807/1560-7917.ES.2019.24.24.1900354. Euro Surveill. 2019. PMID: 31213219 Free PMC article. No abstract available.
References
-
- Anagrius, C. [amp ] Loré, B. (2008). Clinical experiences of new variant Chlamydia trachomatis in a Swedish high prevalence county. In 24th International Union against Sexually Transmitted Infections – Europe conference, Milan, abstract FOC4.12.
-
- Berriman, M. [amp ] Rutherford, K. (2003). Viewing and annotating sequence data with Artemis. Brief Bioinform 4, 124–132. - PubMed
-
- Bjartling, C., Osser, S., Johnson, A. [amp ] Persson, K. (2009). Clinical manifestations and epidemiology of the new genetic variant of Chlamydia trachomatis. Sex Transm Dis 36, 529–535. - PubMed
-
- Caldwell, H. D., Wood, H., Crane, D., Bailey, R., Jones, R. B., Mabey, D., Maclean, I., Mohammed, Z., Peeling, R. [amp ] other authors (2003). Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J Clin Invest 111, 1757–1769. - PMC - PubMed
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

