Predominant occupation of the class I MHC molecule H-2Kwm7 with a single self-peptide suggests a mechanism for its diabetes-protective effect
- PMID: 20093428
- PMCID: PMC2829095
- DOI: 10.1093/intimm/dxp127
Predominant occupation of the class I MHC molecule H-2Kwm7 with a single self-peptide suggests a mechanism for its diabetes-protective effect
Abstract
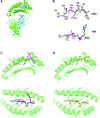
Type 1 diabetes (T1D) is an autoimmune disease characterized by T cell-mediated destruction of insulin-producing pancreatic beta cells. In both humans and the non-obese diabetic (NOD) mouse model of T1D, class II MHC alleles are the primary determinant of disease susceptibility. However, class I MHC genes also influence risk. These findings are consistent with the requirement for both CD4(+) and CD8(+) T cells in the pathogenesis of T1D. Although a large body of work has permitted the identification of multiple mechanisms to explain the diabetes-protective effect of particular class II MHC alleles, studies examining the protective influence of class I alleles are lacking. Here, we explored this question by performing biochemical and structural analyses of the murine class I MHC molecule H-2K(wm7), which exerts a diabetes-protective effect in NOD mice. We have found that H-2K(wm7) molecules are predominantly occupied by the single self-peptide VNDIFERI, derived from the ubiquitous protein histone H2B. This unexpected finding suggests that the inability of H-2K(wm7) to support T1D development could be due, at least in part, to the failure of peptides from critical beta-cell antigens to adequately compete for binding and be presented to T cells. Predominant presentation of a single peptide would also be expected to influence T-cell selection, potentially leading to a reduced ability to select a diabetogenic CD8(+) T-cell repertoire. The report that one of the predominant peptides bound by T1D-protective HLA-A*31 is histone derived suggests the potential translation of our findings to human diabetes-protective class I MHC molecules.
Figures



Similar articles
-
Proteomic identification of MHC class I-associated peptidome derived from non-obese diabetic mouse thymus and pancreas.J Proteomics. 2023 Jan 6;270:104746. doi: 10.1016/j.jprot.2022.104746. Epub 2022 Oct 7. J Proteomics. 2023. PMID: 36210013
-
MHC class II molecules play a role in the selection of autoreactive class I-restricted CD8 T cells that are essential contributors to type 1 diabetes development in nonobese diabetic mice.J Immunol. 2004 Jan 15;172(2):871-9. doi: 10.4049/jimmunol.172.2.871. J Immunol. 2004. PMID: 14707058
-
Self peptides isolated from MHC glycoproteins of non-obese diabetic mice.J Immunol. 1994 Mar 1;152(5):2279-88. J Immunol. 1994. PMID: 8133041
-
Bridging Mice to Men: Using HLA Transgenic Mice to Enhance the Future Prediction and Prevention of Autoimmune Type 1 Diabetes in Humans.Methods Mol Biol. 2016;1438:137-51. doi: 10.1007/978-1-4939-3661-8_9. Methods Mol Biol. 2016. PMID: 27150089 Review.
-
The good turned ugly: immunopathogenic basis for diabetogenic CD8+ T cells in NOD mice.Immunol Rev. 2005 Apr;204:250-63. doi: 10.1111/j.0105-2896.2005.00244.x. Immunol Rev. 2005. PMID: 15790363 Review.
Cited by
-
Characterization of the peptide binding specificity of the HLA class I alleles B*38:01 and B*39:06.Immunogenetics. 2016 Mar;68(3):231-6. doi: 10.1007/s00251-015-0898-2. Epub 2016 Jan 11. Immunogenetics. 2016. PMID: 26754738 Free PMC article.
-
Use of nonobese diabetic mice to understand human type 1 diabetes.Endocrinol Metab Clin North Am. 2010 Sep;39(3):541-61. doi: 10.1016/j.ecl.2010.05.001. Epub 2010 Jul 8. Endocrinol Metab Clin North Am. 2010. PMID: 20723819 Free PMC article. Review.
References
-
- Gianani R, Eisenbarth GS. The stages of type 1A diabetes: 2005. Immunol. Rev. 2005;204:232. - PubMed
-
- Rewers M, Bugawan TL, Norris JM, et al. Newborn screening for HLA markers associated with IDDM: diabetes autoimmunity study in the young (DAISY) Diabetologia. 1996;39:807. - PubMed
-
- Fennessy M, Metcalfe K, Hitman GA, et al. A gene in the HLA class I region contributes to susceptibility to IDDM in the Finnish population. Childhood Diabetes in Finland (DiMe) Study Group. Diabetologia. 1994;37:937. - PubMed
-
- Nakanishi K, Kobayashi T, Murase T, Naruse T, Nose Y, Inoko H. Human leukocyte antigen-A24 and -DQA1*0301 in Japanese insulin-dependent diabetes mellitus: independent contributions to susceptibility to the disease and additive contributions to acceleration of β-cell destruction. J. Clin. Endocrinol. Metab. 1999;84:3721. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

