c-Myb binds MLL through menin in human leukemia cells and is an important driver of MLL-associated leukemogenesis
- PMID: 20093773
- PMCID: PMC2810070
- DOI: 10.1172/JCI38030
c-Myb binds MLL through menin in human leukemia cells and is an important driver of MLL-associated leukemogenesis
Abstract
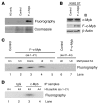
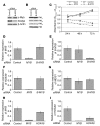
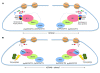
Mixed-lineage leukemia (MLL) is a proto-oncogene frequently involved in chromosomal translocations associated with acute leukemia. These chromosomal translocations commonly result in MLL fusion proteins that dysregulate transcription. Recent data suggest that the MYB proto-oncogene, which is an important regulator of hematopoietic cell development, has a role in leukemogenesis driven by the MLL-ENL fusion protein, but exactly how is unclear. Here we have demonstrated that c-Myb is recruited to the MLL histone methyl transferase complex by menin, a protein important for MLL-associated leukemic transformation, and that it contributes substantially to MLL-mediated methylation of histone H3 at lysine 4 (H3K4). Silencing MYB in human leukemic cell lines and primary patient material evoked a global decrease in H3K4 methylation, an unexpected decrease in HOXA9 and MEIS1 gene expression, and decreased MLL and menin occupancy in the HOXA9 gene locus. This decreased occupancy was associated with a diminished ability of an MLL-ENL fusion protein to transform normal mouse hematopoietic cells. Previous studies have shown that MYB expression is regulated by Hoxa9 and Meis1, indicating the existence of an autoregulatory feedback loop. The finding that c-Myb has the ability to direct epigenetic marks, along with its participation in an autoregulatory feedback loop with genes known to transform hematopoietic cells, lends mechanistic and translationally relevant insight into its role in MLL-associated leukemogenesis.
Figures

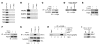

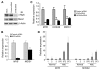

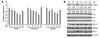
References
-
- Kastan MB, Slamon DJ, Civin CI. Expression of protooncogene c-myb in normal human hematopoietic cells. . Blood. 1989;73(6):1444–1451. - PubMed

