Lessons from the lysozyme of phage T4
- PMID: 20095051
- PMCID: PMC2867005
- DOI: 10.1002/pro.344
Lessons from the lysozyme of phage T4
Abstract
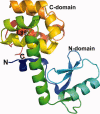
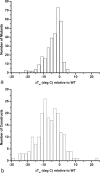
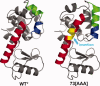
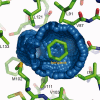
An overview is presented of some of the major insights that have come from studies of the structure, stability, and folding of T4 phage lysozyme. A major purpose of this review is to provide the reader with a complete tabulation of all of the variants that have been characterized, including melting temperatures, crystallographic data, Protein Data Bank access codes, and references to the original literature. The greatest increase in melting temperature (T(m)) for any point mutant is 5.1 degrees C for the mutant Ser 117 --> Val. This is achieved in part not only by hydrophobic stabilization but also by eliminating an unusually short hydrogen bond of 2.48 A that apparently has an unfavorable van der Waals contact. Increases in T(m) of more than 3-4 degrees C for point mutants are rare, whereas several different types of destabilizing substitutions decrease T(m) by 20 degrees C or thereabouts. The energetic cost of cavity creation and its relation to the hydrophobic effect, derived from early studies of "large-to-small" mutants in the core of T4 lysozyme, has recently been strongly supported by related studies of the intrinsic membrane protein bacteriorhodopsin. The L99A cavity in the C-terminal domain of the protein, which readily binds benzene and many other ligands, has been the subject of extensive study. Crystallographic evidence, together with recent NMR analysis, suggest that these ligands are admitted by a conformational change involving Helix F and its neighbors. A total of 43 nonisomorphous crystal forms of different monomeric lysozyme mutants were obtained plus three more for synthetically-engineered dimers. Among the 43 space groups, P2(1)2(1)2(1) and P2(1) were observed most frequently, consistent with the prediction of Wukovitz and Yeates.
Figures

Similar articles
-
The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.Protein Sci. 1998 Jan;7(1):158-77. doi: 10.1002/pro.5560070117. Protein Sci. 1998. PMID: 9514271 Free PMC article.
-
Structural analysis of a non-contiguous second-site revertant in T4 lysozyme shows that increasing the rigidity of a protein can enhance its stability.J Mol Biol. 1999 Oct 8;292(5):1111-20. doi: 10.1006/jmbi.1999.3102. J Mol Biol. 1999. PMID: 10512706
-
Multiple alanine replacements within alpha-helix 126-134 of T4 lysozyme have independent, additive effects on both structure and stability.Protein Sci. 1992 Jun;1(6):761-76. doi: 10.1002/pro.5560010608. Protein Sci. 1992. PMID: 1304917 Free PMC article.
-
Structural and genetic analysis of the folding and function of T4 lysozyme.FASEB J. 1996 Jan;10(1):35-41. doi: 10.1096/fasebj.10.1.8566545. FASEB J. 1996. PMID: 8566545 Review.
-
Studies on protein stability with T4 lysozyme.Adv Protein Chem. 1995;46:249-78. doi: 10.1016/s0065-3233(08)60337-x. Adv Protein Chem. 1995. PMID: 7771320 Review.
Cited by
-
Crystal structure of Bax bound to the BH3 peptide of Bim identifies important contacts for interaction.Cell Death Dis. 2015 Jul 9;6(7):e1809. doi: 10.1038/cddis.2015.141. Cell Death Dis. 2015. PMID: 26158515 Free PMC article.
-
The role of adjuvant in mediating antigen structure and stability.J Pharm Sci. 2012 Apr;101(4):1391-9. doi: 10.1002/jps.23039. Epub 2011 Dec 28. J Pharm Sci. 2012. PMID: 22213631 Free PMC article.
-
Addressing Intersite Coupling Unlocks Large Combinatorial Chemical Spaces for Alchemical Free Energy Methods.J Chem Theory Comput. 2022 Apr 12;18(4):2114-2123. doi: 10.1021/acs.jctc.1c00948. Epub 2022 Mar 7. J Chem Theory Comput. 2022. PMID: 35255214 Free PMC article.
-
Peripatetic proteins.Protein Sci. 2010 Jul;19(7):1279-80. doi: 10.1002/pro.422. Protein Sci. 2010. PMID: 20499369 Free PMC article. No abstract available.
-
Escape of a Small Molecule from Inside T4 Lysozyme by Multiple Pathways.Biophys J. 2018 Mar 13;114(5):1058-1066. doi: 10.1016/j.bpj.2018.01.014. Biophys J. 2018. PMID: 29539393 Free PMC article.
References
-
- Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi H-J, Yao X-J, Weis WI, Stevens RC, Kobilka BK. GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science. 2007;318:1266–1273. - PubMed
-
- Tokuriki N, Tawfik DS. Stability effects of mutations and protein evolvability. Curr Opin Struct Biol. 2009;19:596–604. - PubMed
-
- Rennell D, Bouvier SE, Hardy LW, Poteete AR. Systematic mutation of bacteriophage T4 lysozyme. J Mol Biol. 1991;222:67–87. - PubMed
-
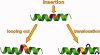
- Heinz DW, Baase WA, Dahlquist FW, Matthews BW. How amino-acid insertions are allowed in an α-helix of T4 lysozyme. Nature. 1993;361:561–564. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

