Brn3a regulates neuronal subtype specification in the trigeminal ganglion by promoting Runx expression during sensory differentiation
- PMID: 20096094
- PMCID: PMC2829025
- DOI: 10.1186/1749-8104-5-3
Brn3a regulates neuronal subtype specification in the trigeminal ganglion by promoting Runx expression during sensory differentiation
Abstract
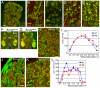
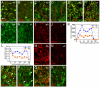
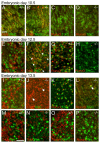
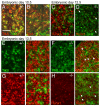
The transcription factor Brn3a, product of the pou4f1 gene, is expressed in most sensory neurons throughout embryogenesis. Prior work has demonstrated a role for Brn3a in the repression of early neurogenic genes; here we describe a second major role for Brn3a in the specification of sensory subtypes in the trigeminal ganglion (TG). Sensory neurons initially co-express multiple Trk-family neurotrophin receptors, but are later marked by the unique expression of TrkA, TrkB or TrkC. Maturation of these sensory subtypes is known to depend on the expression of Runx transcription factors. Newborn Brn3a knockout mice fail to express TrkC, which is associated in the TG with mechanoreceptors, plus a set of functional genes associated with nociceptor subtypes. In embryonic Brn3a-/- ganglia, the normal expression of Runx3 is never initiated in TrkC+ neurons, and Runx1 expression is greatly attenuated in TrkA+ nociceptors. These changes are accompanied by expanded expression of TrkB in neurons that abnormally express multiple Trks, followed by the loss of TrkC and TrkA expression. In transgenic embryos expressing a Brn3a-VP16 dominant transactivator, Runx3 mRNA expression is increased, suggesting that it is a direct regulatory target of Brn3a. Chromatin immunoprecipitation confirms that Brn3a binds in vivo to a conserved upstream enhancer element within histone H3-acetylated chromatin in the Runx3 locus. Together these data show that Brn3a acts upstream of the Runx factors, which then repress TrkB expression to allow establishment of the non-overlapping Trk receptor profiles and correct terminally differentiated phenotypes.
Figures

Similar articles
-
Runx3, Brn3a and Isl1 interplay orchestrates the transcriptional program in the early stages of proprioceptive neuron development.PLoS Genet. 2024 Dec 23;20(12):e1011401. doi: 10.1371/journal.pgen.1011401. eCollection 2024 Dec. PLoS Genet. 2024. PMID: 39715266 Free PMC article.
-
Brn3a/Pou4f1 regulates dorsal root ganglion sensory neuron specification and axonal projection into the spinal cord.Dev Biol. 2012 Apr 15;364(2):114-27. doi: 10.1016/j.ydbio.2012.01.021. Epub 2012 Feb 3. Dev Biol. 2012. PMID: 22326227 Free PMC article.
-
Runx3 is required for the specification of TrkC-expressing mechanoreceptive trigeminal ganglion neurons.Mol Cell Neurosci. 2010 Mar;43(3):296-307. doi: 10.1016/j.mcn.2009.12.003. Epub 2009 Dec 23. Mol Cell Neurosci. 2010. PMID: 20034568
-
Brn3a regulates the transition from neurogenesis to terminal differentiation and represses non-neural gene expression in the trigeminal ganglion.Dev Dyn. 2009 Dec;238(12):3065-79. doi: 10.1002/dvdy.22145. Dev Dyn. 2009. PMID: 19877281 Free PMC article.
-
Transcriptional regulation of Trk family neurotrophin receptors.Cell Mol Life Sci. 2007 Mar;64(5):522-32. doi: 10.1007/s00018-006-6328-8. Cell Mol Life Sci. 2007. PMID: 17192812 Free PMC article. Review.
Cited by
-
Molecular characterization of the mouse superior lateral parabrachial nucleus through expression of the transcription factor Runx1.PLoS One. 2010 Nov 11;5(11):e13944. doi: 10.1371/journal.pone.0013944. PLoS One. 2010. PMID: 21085653 Free PMC article.
-
Generation of Otic Sensory Neurons from Mouse Embryonic Stem Cells in 3D Culture.Front Cell Neurosci. 2017 Dec 19;11:409. doi: 10.3389/fncel.2017.00409. eCollection 2017. Front Cell Neurosci. 2017. PMID: 29311837 Free PMC article.
-
Pou4f1 and pou4f2 are dispensable for the long-term survival of adult retinal ganglion cells in mice.PLoS One. 2014 Apr 15;9(4):e94173. doi: 10.1371/journal.pone.0094173. eCollection 2014. PLoS One. 2014. PMID: 24736625 Free PMC article.
-
The specification and wiring of mammalian cutaneous low-threshold mechanoreceptors.Wiley Interdiscip Rev Dev Biol. 2016 May-Jun;5(3):389-404. doi: 10.1002/wdev.229. Epub 2016 Mar 17. Wiley Interdiscip Rev Dev Biol. 2016. PMID: 26992078 Free PMC article. Review.
-
Developing a sense of touch.Development. 2017 Nov 15;144(22):4078-4090. doi: 10.1242/dev.120402. Development. 2017. PMID: 29138290 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

