Regulation of UGT1A1 and HNF1 transcription factor gene expression by DNA methylation in colon cancer cells
- PMID: 20096102
- PMCID: PMC2835698
- DOI: 10.1186/1471-2199-11-9
Regulation of UGT1A1 and HNF1 transcription factor gene expression by DNA methylation in colon cancer cells
Abstract
Background: UDP-glucuronosyltransferase 1A1 (UGT1A1) is a pivotal enzyme involved in metabolism of SN-38, the active metabolite of irinotecan commonly used to treat metastatic colorectal cancer. We previously demonstrated aberrant methylation of specific CpG dinucleotides in UGT1A1-negative cells, and revealed that methylation state of the UGT1A1 5'-flanking sequence is negatively correlated with gene transcription. Interestingly, one of these CpG dinucleotides (CpG -4) is found close to a HNF1 response element (HRE), known to be involved in activation of UGT1A1 gene expression, and within an upstream stimulating factor (USF) binding site.
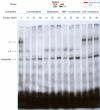
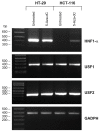
Results: Gel retardation assays revealed that methylation of CpG-4 directly affect the interaction of USF1/2 with its cognate sequence without altering the binding for HNF1-alpha. Luciferase assays sustained a role for USF1/2 and HNF1-alpha in UGT1A1 regulation in colon cancer cells. Based on the differential expression profiles of HNF1A gene in colon cell lines, we also assessed whether methylation affects its expression. In agreement with the presence of CpG islands in the HNF1A promoter, treatments of UGT1A1-negative HCT116 colon cancer cells with a DNA methyltransferase inhibitor restore HNF1A gene expression, as observed for UGT1A1.
Conclusions: This study reveals that basal UGT1A1 expression in colon cells is positively regulated by HNF1-alpha and USF, and negatively regulated by DNA methylation. Besides, DNA methylation of HNF1A could also play an important role in regulating additional cellular drug metabolism and transporter pathways. This process may contribute to determine local inactivation of drugs such as the anticancer agent SN-38 by glucuronidation and define tumoral response.
Figures



Similar articles
-
Irinotecan inactivation is modulated by epigenetic silencing of UGT1A1 in colon cancer.Clin Cancer Res. 2006 Mar 15;12(6):1850-8. doi: 10.1158/1078-0432.CCR-05-2130. Clin Cancer Res. 2006. PMID: 16551870
-
Evidence for regulation of UDP-glucuronosyltransferase (UGT) 1A1 protein expression and activity via DNA methylation in healthy human livers.J Pharm Pharmacol. 2013 Jun;65(6):874-83. doi: 10.1111/jphp.12053. Epub 2013 Mar 21. J Pharm Pharmacol. 2013. PMID: 23647681 Free PMC article.
-
Activation of the mouse TATA-less and human TATA-containing UDP-glucuronosyltransferase 1A1 promoters by hepatocyte nuclear factor 1.Mol Pharmacol. 1999 Sep;56(3):526-36. doi: 10.1124/mol.56.3.526. Mol Pharmacol. 1999. PMID: 10462540
-
Nuclear factor κB down-regulates human UDP-glucuronosyltransferase 1A1: a novel mechanism involved in inflammation-associated hyperbilirubinaemia.Biochem J. 2013 Feb 1;449(3):761-70. doi: 10.1042/BJ20121055. Biochem J. 2013. PMID: 23130636
-
Proximal HNF1 element is essential for the induction of human UDP-glucuronosyltransferase 1A1 by glucocorticoid receptor.Biochem Pharmacol. 2006 Feb 28;71(5):693-701. doi: 10.1016/j.bcp.2005.11.014. Epub 2005 Dec 19. Biochem Pharmacol. 2006. PMID: 16360646
Cited by
-
Multiomics dynamic learning enables personalized diagnosis and prognosis for pancancer and cancer subtypes.Brief Bioinform. 2023 Sep 22;24(6):bbad378. doi: 10.1093/bib/bbad378. Brief Bioinform. 2023. PMID: 37889117 Free PMC article.
-
Emerging targets in cancer drug resistance.Cancer Drug Resist. 2019 Jun 19;2(2):161-177. doi: 10.20517/cdr.2018.27. eCollection 2019. Cancer Drug Resist. 2019. PMID: 35582722 Free PMC article. Review.
-
Cancer stem cells and chemoresistance: The smartest survives the raid.Pharmacol Ther. 2016 Apr;160:145-58. doi: 10.1016/j.pharmthera.2016.02.008. Epub 2016 Feb 17. Pharmacol Ther. 2016. PMID: 26899500 Free PMC article. Review.
-
Upstream stimulatory factor 1 activates GATA5 expression through an E-box motif.Biochem J. 2012 Aug 15;446(1):89-98. doi: 10.1042/BJ20111942. Biochem J. 2012. PMID: 22625849 Free PMC article.
-
Genome-Wide Discriminatory Information Patterns of Cytosine DNA Methylation.Int J Mol Sci. 2016 Jun 17;17(6):938. doi: 10.3390/ijms17060938. Int J Mol Sci. 2016. PMID: 27322251 Free PMC article.
References
-
- Douillard JY, Cunningham D, Roth AD, Navarro M, James RD, Karasek P, Jandik P, Iveson T, Carmichael J, Alakl M. Irinotecan combined with fluorouracil compared with fluorouracil alone as first-line treatment for metastatic colorectal cancer: a multicentre randomised trial. Lancet. 2000;355(9209):1041–1047. doi: 10.1016/S0140-6736(00)02034-1. - DOI - PubMed
-
- Rougier P, Van Cutsem E, Bajetta E, Niederle N, Possinger K, Labianca R, Navarro M, Morant R, Bleiberg H, Wils J. Randomised trial of irinotecan versus fluorouracil by continuous infusion after fluorouracil failure in patients with metastatic colorectal cancer. Lancet. 1998;352(9138):1407–1412. doi: 10.1016/S0140-6736(98)03085-2. - DOI - PubMed
-
- Kawato Y, Aonuma M, Hirota Y, Kuga H, Sato K. Intracellular roles of SN-38, a metabolite of the camptothecin derivative CPT-11, in the antitumor effect of CPT-11. Cancer Res. 1991;51(16):4187–4191. - PubMed
-
- Haaz MC, Rivory L, Riche C, Vernillet L, Robert J. Metabolism of irinotecan (CPT-11) by human hepatic microsomes: participation of cytochrome P-450 3A and drug interactions. Cancer Res. 1998;58(3):468–472. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

