Expression profiling of S. pombe acetyltransferase mutants identifies redundant pathways of gene regulation
- PMID: 20096118
- PMCID: PMC2823694
- DOI: 10.1186/1471-2164-11-59
Expression profiling of S. pombe acetyltransferase mutants identifies redundant pathways of gene regulation
Abstract
Background: Histone acetyltransferase enzymes (HATs) are implicated in regulation of transcription. HATs from different families may overlap in target and substrate specificity.
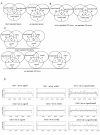
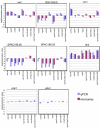
Results: We isolated the elp3+ gene encoding the histone acetyltransferase subunit of the Elongator complex in fission yeast and characterized the phenotype of an Deltaelp3 mutant. We examined genetic interactions between Deltaelp3 and two other HAT mutants, Deltamst2 and Deltagcn5 and used whole genome microarray analysis to analyze their effects on gene expression.
Conclusions: Comparison of phenotypes and expression profiles in single, double and triple mutants indicate that these HAT enzymes have overlapping functions. Consistent with this, overlapping specificity in histone H3 acetylation is observed. However, there is no evidence for overlap with another HAT enzyme, encoded by the essential mst1+ gene.
Figures


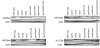
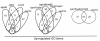
Similar articles
-
The role of specific HAT-HDAC interactions in transcriptional elongation.Cell Cycle. 2010 Feb 1;9(3):467-71. doi: 10.4161/cc.9.3.10543. Cell Cycle. 2010. PMID: 20081370
-
Overlapping roles for the histone acetyltransferase activities of SAGA and elongator in vivo.EMBO J. 2000 Jun 15;19(12):3060-8. doi: 10.1093/emboj/19.12.3060. EMBO J. 2000. PMID: 10856249 Free PMC article.
-
The histone acetyltransferase Mst2 sustains the biological control potential of a fungal insect pathogen through transcriptional regulation.Appl Microbiol Biotechnol. 2018 Feb;102(3):1343-1355. doi: 10.1007/s00253-017-8703-9. Epub 2017 Dec 23. Appl Microbiol Biotechnol. 2018. PMID: 29275430
-
The fission yeast inhibitor of growth (ING) protein Png1p functions in response to DNA damage.J Biol Chem. 2010 May 21;285(21):15786-93. doi: 10.1074/jbc.M110.101832. Epub 2010 Mar 18. J Biol Chem. 2010. PMID: 20299455 Free PMC article.
-
The SAGA histone acetyltransferase module targets SMC5/6 to specific genes.Epigenetics Chromatin. 2023 Feb 16;16(1):6. doi: 10.1186/s13072-023-00480-z. Epigenetics Chromatin. 2023. PMID: 36793083 Free PMC article.
Cited by
-
Gcn5 facilitates Pol II progression, rather than recruitment to nucleosome-depleted stress promoters, in Schizosaccharomyces pombe.Nucleic Acids Res. 2011 Aug;39(15):6369-79. doi: 10.1093/nar/gkr255. Epub 2011 Apr 22. Nucleic Acids Res. 2011. PMID: 21515633 Free PMC article.
-
The SAGA histone acetyltransferase complex regulates leucine uptake through the Agp3 permease in fission yeast.J Biol Chem. 2012 Nov 2;287(45):38158-67. doi: 10.1074/jbc.M112.411165. Epub 2012 Sep 18. J Biol Chem. 2012. PMID: 22992726 Free PMC article.
-
The histone code of the fungal genus Aspergillus uncovered by evolutionary and proteomic analyses.Microb Genom. 2022 Sep;8(9):mgen000856. doi: 10.1099/mgen.0.000856. Microb Genom. 2022. PMID: 36129736 Free PMC article.
-
Anti-silencing factor Epe1 associates with SAGA to regulate transcription within heterochromatin.Genes Dev. 2019 Jan 1;33(1-2):116-126. doi: 10.1101/gad.318030.118. Epub 2018 Dec 20. Genes Dev. 2019. PMID: 30573453 Free PMC article.
-
Elimination of a specific histone H3K14 acetyltransferase complex bypasses the RNAi pathway to regulate pericentric heterochromatin functions.Genes Dev. 2011 Feb 1;25(3):214-9. doi: 10.1101/gad.1993611. Genes Dev. 2011. PMID: 21289066 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

