Many families of C. elegans microRNAs are not essential for development or viability
- PMID: 20096582
- PMCID: PMC2844791
- DOI: 10.1016/j.cub.2009.12.051
Many families of C. elegans microRNAs are not essential for development or viability
Abstract
MicroRNAs (miRNAs) are approximately 23 nt regulatory RNAs that posttranscriptionally inhibit the functions of protein-coding mRNAs. We previously found that most C. elegans miRNAs are individually not essential for development or viability and proposed that paralogous miRNAs might often function redundantly. To test this hypothesis, we generated mutant C. elegans strains that each lack multiple or all members of one of 15 miRNA families. Mutants for 12 of these families did not display strong synthetic abnormalities, suggesting that these miRNA families have subtle roles during development. By contrast, mutants deleted for all members of the mir-35 or mir-51 families died as embryos or early larvae, and mutants deleted for four members of the mir-58 family showed defects in locomotion, body size, and egg laying and an inability to form dauer larvae. Our findings indicate that the regulatory functions of most individual miRNAs and most individual families of miRNAs related in sequence are not critical for development or viability. Conversely, because in some cases miRNA family members act redundantly, our findings emphasize the importance of determining miRNA function in the absence of miRNAs related in sequence.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures
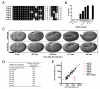

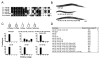
Comment in
-
MicroRNAs: genetically sensitized worms reveal new secrets.Curr Biol. 2010 Jul 27;20(14):R598-600. doi: 10.1016/j.cub.2010.05.054. Curr Biol. 2010. PMID: 20656201
References
-
- Ruby JG, Jan C, Player C, Axtell MJ, Lee W, Nusbaum C, Ge H, Bartel DP. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. - PubMed
-
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

