Parp1 activation in mouse embryonic fibroblasts promotes Pol beta-dependent cellular hypersensitivity to alkylation damage
- PMID: 20096707
- PMCID: PMC2834876
- DOI: 10.1016/j.mrfmmm.2010.01.016
Parp1 activation in mouse embryonic fibroblasts promotes Pol beta-dependent cellular hypersensitivity to alkylation damage
Abstract
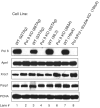
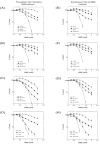
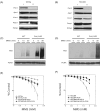
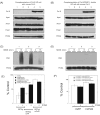
Alkylating agents induce cell death in wild-type (WT) mouse embryonic fibroblasts (MEFs) by multiple mechanisms, including apoptosis, autophagy and necrosis. DNA polymerase beta (Pol beta) knockout (KO) MEFs are hypersensitive to the cytotoxic effect of alkylating agents, as compared to WT MEFs. To test the hypothesis that Parp1 is preferentially activated by methyl methanesulfonate (MMS) exposure of Pol beta KO MEFs, we have examined the relationship between Pol beta expression, Parp1 activation and cell survival following MMS exposure in a series of WT and Pol beta deficient MEF cell lines. Consistent with our hypothesis, we observed elevated Parp1 activation in Pol beta KO MEFs as compared to matched WT MEFs. Both the MMS-induced activation of Parp1 and the MMS-induced cytotoxicity of Pol beta KO MEFs are attenuated by pre-treatment with the Parp1/Parp2 inhibitor PJ34. Further, elevated Parp1 activation is observed following knockdown (KD) of endogenous Pol beta, as compared to WT cells. Pol beta KD MEFs are hypersensitive to MMS and both the MMS-induced hypersensitivity and Parp1 activation is prevented by pre-treatment with PJ34. In addition, the MMS-induced cellular sensitivity of Pol beta KO MEFs is reversed when Parp1 is also deleted (Pol beta/Parp1 double KO MEFs) and we observe no MMS sensitivity differential between Pol beta/Parp1 double KO MEFs and those that express recombinant mouse Pol beta. These studies suggest that Parp1 may function as a sensor of BER to initiate cell death when BER is aborted or fails. Parp1 may therefore function in BER as a tumor suppressor by initiating cell death and preventing the accumulation of cells with chromosomal damage due to a BER defect.
Copyright 2010 Elsevier B.V. All rights reserved.
Conflict of interest statement
Figures

Similar articles
-
Preventing oxidation of cellular XRCC1 affects PARP-mediated DNA damage responses.DNA Repair (Amst). 2013 Sep;12(9):774-85. doi: 10.1016/j.dnarep.2013.06.004. Epub 2013 Jul 18. DNA Repair (Amst). 2013. PMID: 23871146 Free PMC article.
-
AP endonuclease knockdown enhances methyl methanesulfonate hypersensitivity of DNA polymerase β knockout mouse embryonic fibroblasts.J Radiat Res. 2015 May;56(3):462-6. doi: 10.1093/jrr/rru125. Epub 2015 Feb 26. J Radiat Res. 2015. PMID: 25724755 Free PMC article.
-
DNA polymerase β-dependent cell survival independent of XRCC1 expression.DNA Repair (Amst). 2015 Feb;26:23-9. doi: 10.1016/j.dnarep.2014.11.008. Epub 2014 Dec 3. DNA Repair (Amst). 2015. PMID: 25541391 Free PMC article.
-
Mammalian DNA beta-polymerase in base excision repair of alkylation damage.Prog Nucleic Acid Res Mol Biol. 2001;68:57-74. doi: 10.1016/s0079-6603(01)68090-5. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11554313 Review.
-
Hypersensitivity phenotypes associated with genetic and synthetic inhibitor-induced base excision repair deficiency.DNA Repair (Amst). 2007 Apr 1;6(4):530-43. doi: 10.1016/j.dnarep.2006.10.016. Epub 2006 Nov 20. DNA Repair (Amst). 2007. PMID: 17113833 Free PMC article. Review.
Cited by
-
The frequency and clinical significance of DNA polymerase beta (POLβ) expression in breast ductal carcinoma in situ (DCIS).Breast Cancer Res Treat. 2021 Nov;190(1):39-51. doi: 10.1007/s10549-021-06357-7. Epub 2021 Aug 18. Breast Cancer Res Treat. 2021. PMID: 34406589 Free PMC article.
-
Genome instability caused by a germline mutation in the human DNA repair gene POLB.PLoS Genet. 2012;8(11):e1003086. doi: 10.1371/journal.pgen.1003086. Epub 2012 Nov 8. PLoS Genet. 2012. PMID: 23144636 Free PMC article. No abstract available.
-
Temporal dynamics of base excision/single-strand break repair protein complex assembly/disassembly are modulated by the PARP/NAD+/SIRT6 axis.Cell Rep. 2021 Nov 2;37(5):109917. doi: 10.1016/j.celrep.2021.109917. Cell Rep. 2021. PMID: 34731617 Free PMC article.
-
Apurinic/apyrimidinic (AP) site recognition by the 5'-dRP/AP lyase in poly(ADP-ribose) polymerase-1 (PARP-1).Proc Natl Acad Sci U S A. 2010 Dec 21;107(51):22090-5. doi: 10.1073/pnas.1009182107. Epub 2010 Dec 2. Proc Natl Acad Sci U S A. 2010. PMID: 21127267 Free PMC article.
-
Transcriptional profiling reveals elevated Sox2 in DNA polymerase ß null mouse embryonic fibroblasts.Am J Cancer Res. 2012;2(6):699-713. doi: 10.1158/1538-7445.am2012-699. Epub 2012 Nov 20. Am J Cancer Res. 2012. PMID: 23226616 Free PMC article.
References
-
- Sobol RW, Horton JK, Kuhn R, Gu H, Singhal RK, Prasad R, Rajewsky K, Wilson SH. Requirement of mammalian DNA polymerase-β in base-excision repair. Nature. 1996;379:183–186. - PubMed
-
- Beard WA, Wilson SH. Structure and mechanism of DNA polymerase Beta. Chem Rev. 2006;106:361–382. - PubMed
-
- Horton JK, Prasad R, Hou E, Wilson SH. Protection against methylation-induced cytotoxicity by DNA polymerase β-dependent long patch base excision repair. Journal of Biological Chemistry. 2000;275:2211–2218. - PubMed
-
- Fortini P, Pascucci B, Parlanti E, Sobol RW, Wilson SH, Dogliotti E. Different DNA polymerases are involved in the short- and long-patch base excision repair in mammalian cells. Biochemistry. 1998;37:3575–3580. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

