Label-free microarray imaging for direct detection of DNA hybridization and single-nucleotide mismatches
- PMID: 20097056
- PMCID: PMC2824047
- DOI: 10.1016/j.bios.2009.12.032
Label-free microarray imaging for direct detection of DNA hybridization and single-nucleotide mismatches
Abstract
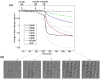
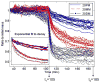
A novel method is proposed for direct detection of DNA hybridization on microarrays. Optical interferometry is used for label-free sensing of biomolecular accumulation on glass surfaces, enabling dynamic detection of interactions. Capabilities of the presented method are demonstrated by high-throughput sensing of solid-phase hybridization of oligonucleotides. Hybridization of surface immobilized probes with 20 base pair-long target oligonucleotides was detected by comparing the label-free microarray images taken before and after hybridization. Through dynamic data acquisition during denaturation by washing the sample with low ionic concentration buffer, melting of duplexes with a single-nucleotide mismatch was distinguished from perfectly matching duplexes with high confidence interval (>97%). The presented technique is simple, robust, and accurate, and eliminates the need of using labels or secondary reagents to monitor the oligonucleotide hybridization.
(c) 2010 Elsevier B.V. All rights reserved.
Figures


References
-
- Cooper MA. Optical biosensors in drug discovery. Nature Reviews Drug Discovery. 2002;1(7):515–528. - PubMed
-
- Cooper MA. Label-free screening of bio-molecular interactions. Analytical And Bioanalytical Chemistry. 2003;377(5):834–842. - PubMed
-
- Cretich M, Pirri G, Damin F, Solinas I, Chiari M. A new polymeric coating for protein microarrays. Analytical Biochemistry. 2004;332(1):67–74. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

