Recruitment to stalled replication forks of the PriA DNA helicase and replisome-loading activities is essential for survival
- PMID: 20097140
- PMCID: PMC2827650
- DOI: 10.1016/j.dnarep.2009.12.009
Recruitment to stalled replication forks of the PriA DNA helicase and replisome-loading activities is essential for survival
Abstract
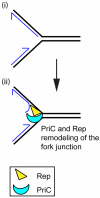
PriA, a 3'-->5' superfamily 2 DNA helicase, acts to remodel stalled replication forks and as a specificity factor for origin-independent assembly of a new replisome at the stalled fork. The ability of PriA to initiate replication at stalled forked structures ensures complete genome replication and helps to protect the cell from illegitimate recombination events. This review focuses on the activities of PriA and its role in replication fork assembly and maintaining genomic integrity.
(c) 2010 Elsevier B.V. All rights reserved.
Figures

References
-
- Allen GC, Jr., Kornberg A. Assembly of the primosome of DNA replication in Escherichia coli. J Biol Chem. 1993;268:19204–9. - PubMed
-
- Arber W. Host-controlled variation. In: Hershey AD, editor. The Bacteriophage Lambda. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1971. pp. 83–96.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

