Resonance assignment and three-dimensional structure determination of a human alpha-defensin, HNP-1, by solid-state NMR
- PMID: 20097206
- PMCID: PMC2873975
- DOI: 10.1016/j.jmb.2010.01.030
Resonance assignment and three-dimensional structure determination of a human alpha-defensin, HNP-1, by solid-state NMR
Abstract
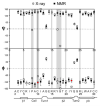
Human alpha-defensins [human neutrophil peptides (HNPs)] are immune defense mini-proteins that act by disrupting microbial cell membranes. Elucidating the three-dimensional (3D) structures of HNPs in lipid membranes is important for understanding their mechanisms of action. Using solid-state NMR (SSNMR), we have determined the 3D structure of HNP-1 in a microcrystalline state outside the lipid membrane, which provides benchmarks for structure determination and comparison with the membrane-bound state. From a suite of two-dimensional and 3D magic-angle spinning experiments, (13)C and (15)N chemical shifts that yielded torsion angle constraints were obtained, while inter-residue distances were obtained to restrain the 3D fold. Together, these constraints led to the first high-resolution SSNMR structure of a human defensin. The SSNMR structure has close similarity to the crystal structures of the HNP family, with the exception of the loop region between the first and second beta-strands. The difference, which is partially validated by direct torsion angle measurements of selected loop residues, suggests possible conformational variation and flexibility of this segment of the protein, which may regulate HNP interaction with the phospholipid membrane of microbial cells.
Figures








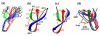

Similar articles
-
The membrane-bound structure and topology of a human α-defensin indicate a dimer pore mechanism for membrane disruption.Biochemistry. 2010 Nov 16;49(45):9770-82. doi: 10.1021/bi101512j. Epub 2010 Oct 26. Biochemistry. 2010. PMID: 20961099 Free PMC article.
-
3D (13)C-(13)C-(13)C correlation NMR for de novo distance determination of solid proteins and application to a human alpha-defensin.J Magn Reson. 2010 Feb;202(2):203-10. doi: 10.1016/j.jmr.2009.11.011. Epub 2009 Nov 18. J Magn Reson. 2010. PMID: 19963419 Free PMC article.
-
NMR studies of defensin antimicrobial peptides. 1. Resonance assignment and secondary structure determination of rabbit NP-2 and human HNP-1.Biochemistry. 1992 Nov 24;31(46):11348-56. doi: 10.1021/bi00161a012. Biochemistry. 1992. PMID: 1445872
-
Solid-State NMR of Membrane Proteins in Lipid Bilayers: To Spin or Not To Spin?Acc Chem Res. 2021 Mar 16;54(6):1430-1439. doi: 10.1021/acs.accounts.0c00670. Epub 2021 Mar 3. Acc Chem Res. 2021. PMID: 33655754 Free PMC article. Review.
-
Recent advances in magic angle spinning solid state NMR of membrane proteins.Prog Nucl Magn Reson Spectrosc. 2014 Oct;82:1-26. doi: 10.1016/j.pnmrs.2014.07.001. Epub 2014 Jul 26. Prog Nucl Magn Reson Spectrosc. 2014. PMID: 25444696 Review.
Cited by
-
Secondary structure in the core of amyloid fibrils formed from human β₂m and its truncated variant ΔN6.J Am Chem Soc. 2014 Apr 30;136(17):6313-25. doi: 10.1021/ja4126092. Epub 2014 Apr 16. J Am Chem Soc. 2014. PMID: 24679070 Free PMC article.
-
Single, double and quadruple alanine substitutions at oligomeric interfaces identify hydrophobicity as the key determinant of human neutrophil alpha defensin HNP1 function.PLoS One. 2013 Nov 13;8(11):e78937. doi: 10.1371/journal.pone.0078937. eCollection 2013. PLoS One. 2013. PMID: 24236072 Free PMC article.
-
Protein fold determined by paramagnetic magic-angle spinning solid-state NMR spectroscopy.Nat Chem. 2012 Mar 18;4(5):410-7. doi: 10.1038/nchem.1299. Nat Chem. 2012. PMID: 22522262 Free PMC article.
-
Sequence conservation, phylogenetic relationships, and expression profiles of nondigestive serine proteases and serine protease homologs in Manduca sexta.Insect Biochem Mol Biol. 2015 Jul;62:51-63. doi: 10.1016/j.ibmb.2014.10.006. Epub 2014 Dec 19. Insect Biochem Mol Biol. 2015. PMID: 25530503 Free PMC article.
-
The Antimicrobial and Antiviral Applications of Cell-Penetrating Peptides.Methods Mol Biol. 2015;1324:223-45. doi: 10.1007/978-1-4939-2806-4_15. Methods Mol Biol. 2015. PMID: 26202273 Free PMC article. Review.
References
-
- Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 2005;6:551–557. - PubMed
-
- Hill CP, Yee J, Selsted ME, Eisenberg D. Crystal structure of defensin HNP-3, an amphiphilic dimer: mechanisms of membrane permeabilization. Science. 1991;251:1481–1485. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

