Evidence for ribosomal frameshifting and a novel overlapping gene in the genomes of insect-specific flaviviruses
- PMID: 20097399
- PMCID: PMC2830293
- DOI: 10.1016/j.virol.2009.12.033
Evidence for ribosomal frameshifting and a novel overlapping gene in the genomes of insect-specific flaviviruses
Abstract
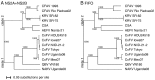
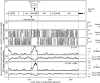
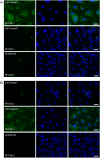
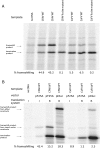
Flaviviruses have a positive-sense, single-stranded RNA genome of approximately 11 kb, encoding a large polyprotein that is cleaved to produce approximately 10 mature proteins. Cell fusing agent virus, Kamiti River virus, Culex flavivirus and several recently discovered flaviviruses have no known vertebrate host and apparently infect only insects. We present compelling bioinformatic evidence for a 253-295 codon overlapping gene (designated fifo) conserved throughout these insect-specific flaviviruses and immunofluorescent detection of its product. Fifo overlaps the NS2A/NS2B coding sequence in the -1/+2 reading frame and is most likely expressed as a trans-frame fusion protein via ribosomal frameshifting at a conserved GGAUUUY slippery heptanucleotide with 3'-adjacent RNA secondary structure (which stimulates efficient frameshifting in vitro). The discovery bears striking parallels to the recently discovered ribosomal frameshifting site in the NS2A coding sequence of the Japanese encephalitis serogroup of flaviviruses and suggests that programmed ribosomal frameshifting may be more widespread in flaviviruses than currently realized.
Copyright 2009 Elsevier Inc. All rights reserved.
Figures





Similar articles
-
Detection of Quang Binh virus from mosquitoes in China.Virus Res. 2014 Feb 13;180:31-8. doi: 10.1016/j.virusres.2013.12.005. Epub 2013 Dec 14. Virus Res. 2014. PMID: 24342141
-
A conserved predicted pseudoknot in the NS2A-encoding sequence of West Nile and Japanese encephalitis flaviviruses suggests NS1' may derive from ribosomal frameshifting.Virol J. 2009 Feb 5;6:14. doi: 10.1186/1743-422X-6-14. Virol J. 2009. PMID: 19196463 Free PMC article.
-
Ribosomal frameshifting into an overlapping gene in the 2B-encoding region of the cardiovirus genome.Proc Natl Acad Sci U S A. 2011 Nov 15;108(46):E1111-9. doi: 10.1073/pnas.1102932108. Epub 2011 Oct 24. Proc Natl Acad Sci U S A. 2011. PMID: 22025686 Free PMC article.
-
Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use.Nucleic Acids Res. 2016 Sep 6;44(15):7007-78. doi: 10.1093/nar/gkw530. Epub 2016 Jul 19. Nucleic Acids Res. 2016. PMID: 27436286 Free PMC article. Review.
-
A review on architecture of the gag-pol ribosomal frameshifting RNA in human immunodeficiency virus: a variability survey of virus genotypes.J Biomol Struct Dyn. 2017 Jun;35(8):1629-1653. doi: 10.1080/07391102.2016.1194231. Epub 2016 Aug 2. J Biomol Struct Dyn. 2017. PMID: 27485859 Review.
Cited by
-
Novel virus discovery and genome reconstruction from field RNA samples reveals highly divergent viruses in dipteran hosts.PLoS One. 2013 Nov 18;8(11):e80720. doi: 10.1371/journal.pone.0080720. eCollection 2013. PLoS One. 2013. PMID: 24260463 Free PMC article.
-
A Novel Virus of Flaviviridae Associated with Sexual Precocity in Macrobrachium rosenbergii.mSystems. 2021 Jun 29;6(3):e0000321. doi: 10.1128/mSystems.00003-21. Epub 2021 Jun 8. mSystems. 2021. PMID: 34100644 Free PMC article.
-
Discovery of Novel Crustacean and Cephalopod Flaviviruses: Insights into the Evolution and Circulation of Flaviviruses between Marine Invertebrate and Vertebrate Hosts.J Virol. 2019 Jun 28;93(14):e00432-19. doi: 10.1128/JVI.00432-19. Print 2019 Jul 15. J Virol. 2019. PMID: 31068424 Free PMC article.
-
An alternative -1/+2 open reading frame exists within viral N(pro)(1-19) region of bovine viral diarrhea virus SD-1.Virus Res. 2012 Jan;163(1):341-51. doi: 10.1016/j.virusres.2011.10.022. Epub 2011 Nov 4. Virus Res. 2012. PMID: 22079882 Free PMC article.
-
Chimeric Zika viruses containing structural protein genes of insect-specific flaviviruses cannot replicate in vertebrate cells due to entry and post-translational restrictions.Virology. 2021 Jul;559:30-39. doi: 10.1016/j.virol.2021.03.014. Epub 2021 Mar 26. Virology. 2021. PMID: 33812340 Free PMC article.
References
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Balmori Melian, E., Hinzman, E., Nagasaki, T., Firth, A.E., Wills, N.M., Nouwens, A.S., Blitvich, B.J., Leung, J., Funk, A., Atkins, J.F., Hall, R., Khromykh, A.A., 2009. NS1′ of flaviviruses in the Japanese encephalitis serogroup is a product of ribosomal frameshifting and plays a role in viral neuro-invasiveness, J. Virol. doi:10.1128/JVI.01979-09. - DOI - PMC - PubMed
-
- Baranov P.V., Gesteland R.F., Atkins J.F. Recoding: translational bifurcations in gene expression. Gene. 2002;286:187–201. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

