Modulation of type I IFN induction by a virulence determinant within the alphavirus nsP1 protein
- PMID: 20097400
- PMCID: PMC2830325
- DOI: 10.1016/j.virol.2009.12.031
Modulation of type I IFN induction by a virulence determinant within the alphavirus nsP1 protein
Abstract
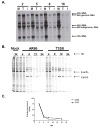
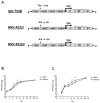
Alphaviruses are mosquito-borne viruses that cause serious human and animal diseases. Previous studies demonstrated that a determinant within the nsP1/nsP2 cleavage domain of the virulent Sindbis AR86 virus played a key role in regulating adult mouse virulence without adversely affecting viral replication. Additional characterization of this determinant demonstrated that a virus with the attenuating mutation induced more type I IFN production both in vivo and in vitro. Interestingly, this phenotype was not specific to the Sindbis AR86 virus, as a similar mutation in a distantly related alphavirus, Ross River Virus (RRV), also led to enhanced IFN induction. This effect was independent of virus-induced host shutoff, since IRF-3 phosphorylation, which occurs independently of de novo host transcription/translation, was induced more robustly in cells infected with the mutant viruses. Altogether, these results demonstrate that critical determinants within the nsP1/nsP2 cleavage domain play an important role in regulating alphavirus-induced IFN responses.
Copyright 2009 Elsevier Inc. All rights reserved.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

